Abstract
Inactivation of the function of tumor suppressor p53 is common in human cancers. In approximately half of human cancers, the tumor suppressor function of p53 is inactivated by deletion or mutation of TP53, the gene encoding p53 protein. In the remaining 50% of human cancers, p53 tumor suppressor function can be effectively inhibited by oncoprotein MDM2 or its homolog MDMX. Since inhibition of p53 by MDM2 or MDMX protein is mediated by their direct interaction with p53, small-molecule inhibitors designed to block the MDM2-p53 or MDMX-p53 protein-protein interaction (MDM2 or MDMX inhibitors) can activate p53 in tumor cells retaining wild-type p53. In the last few years, several classes of potent, selective, and efficacious small molecule MDM2 inhibitors have been designed and developed, and six such compounds are being evaluated in clinical trials as new anticancer drugs. Additionally, non-peptide, small-molecule MDMX inhibitors have been reported. We review herein the design and development of potent small-molecule MDM2 and MDMX inhibitors.
Citation: Zhao Y, Bernard D and Wang S. Small Molecule inhibitors of MDM2-p53 and MDMX-p53 interaction as new cancer therapeutics. BioDiscovery2013; 8: 4; DOI: 10.7750/BioDiscovery.2013.8.4
Copyright: © 2013 Zhao et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, provided the original authors and source are credited.
Received: May 1, 2013; Accepted: July 2, 2013; Available online /Published: July 8, 2013
Keywords: protein-protein interaction, p53, MDM2, MDMX, structure-based design, small-molecule inhibitors of MDM2, small-molecule inhibitors of MDMX
*Corresponding Author: S. Wang, e-mail: shaomeng@med.umich.edu
Conflict of Interests: Drs. Shaomeng Wang and Yujun Zhao are inventors for the MDM2 inhibitors discovered at the University of Michigan and developed by Sanofi and receive royalty from the University of Michigan.
Introduction
p53 and its primary cellular inhibitor MDM2
The transcription factor p53 plays an important role in regulation of cell cycle, apoptosis, DNA repair, senescence, angiogenesis, cellular metabolism, and innate immunity [1-3]. The p53 protein was identified in 1979 [4-6], and TP53, the gene encoding p53, was cloned in 1983 [7]. Although initially thought of as an oncogene, wild-type p53 is in fact a powerful tumor suppressor. However, in nearly 50% of human cancers, the tumor suppressor function of p53 is inactivated by mutation or deletion of its gene [8]. In the other 50% of human cancers, p53 retains wild-type status but its tumor suppressor function can be compromised by multiple mechanisms, and one major inhibitory mechanism is mediated by its primary cellular inhibitor, the murine double minute 2 protein (MDM2; known as HDM2 in human). MDM2 was initially discovered by its overexpression in a spontaneously transformed mouse cell line [8-12]. MDM2 and p53 mutually regulate each other through an auto-regulatory feedback loop [13, 14]. As a transcription factor, p53 increases the expression level of the MDM2 gene upon activation. In turn, MDM2 protein binds directly to p53 and inhibits p53 activity through multiple mechanisms: (1) MDM2 inhibits the transactivation function of p53, (2) MDM2 facilitates export of p53 out of the cell nucleus, making p53 inaccessible to targeted DNA for transcription and (3) MDM2 promotes degradation of p53 by ubiquitination through its E3 ubiquitin ligase activity [13-15]. The physiological relevance of this auto-regulatory loop has been clearly demonstrated by genetic evidence that the embryonic lethality of MDM2-null mice can be rescued only upon concurrent deletion of TP53 [16, 17].
Deregulation of the p53 and MDM2 auto-regulatory loop causes malignant transformation of normal cells. For example, overexpression of MDM2 neutralizes p53 tumor suppressor function in cell differentiation, provides cells with a growth advantage and promotes tumorigenesis in mice [18]. MDM2 overexpression can be promoted by amplification of its gene, single nucleotide polymorphism at nucleotide 309 (SNP309) in its gene promoter, enhanced transcription, or increased translation [19-25]. Overexpression of MDM2 in human tumors correlates with poor clinical prognosis and poor treatment response to current cancer therapies [19, 25-27]. Amplification of MDM2 gene has been found in 7% of human cancers in an analysis of 28 different types of human cancers comprising nearly 4,000 human tumor samples, and amplification of MDM2 and mutations of p53 are mutually exclusive [23]. Mice lacking p53 develop normally but are prone to develop a variety of tumors [28]. Collectively, these data support the notion that MDM2 is a major, though not the only, negative regulator of p53 function.
The role of MDMX in negative regulation of p53
In addition to MDM2, MDMX (also known as MDM4, or as HDMX or HDM4 in human) is another negative regulator of p53 function. MDMX was initially identified in 1996 as a homolog of the p53-binding protein, MDM2, in humans [29, 30]. Structurally, both MDMX and MDM2 have an N-terminal p53 binding domain, an acidic domain, a zinc finger domain, and a C-terminal RING domain. These two proteins share high sequence homology in their p53-binding domains and RING finger domains.Although MDMX has high sequence homology to MDM2, deletion of the MDMX gene in mice is also embryonically lethal and this phenotype can be rescued upon p53 deactivation [31], similar to the phenotype found in MDM2-deficient mice, suggesting that MDM2 and MDMX don’t have a redundant role in regulation of p53 [16, 17].
Although both MDM2 and MDMX inhibit the transcription activity of p53, MDM2 and MDMX have significant differences in their mechanisms of p53 regulation. First, MDM2 has intrinsic E3 ubiquitin ligase activity whereas MDMX does not [32, 33]. In addition, MDMX and MDM2 can form heterodimers mediated by the RING domain in MDM [34, 35] and this MDM2/MDMX complex induces polyubiquitination of p53, whereas MDM2 alone is primarily responsible for monoubiquitination of p53 [36]. Disruption of MDM2/MDMX heterocomplex is embryonically lethal in mice, suggesting that MDM2 and MDMX cooperate to inhibit the activity of p53 [37, 38]. Finally, in contrast to MDM2, MDMX is not under transcriptional control of p53 [39]. Hence, MDM2 and MDMX are important and non-redundant regulators of p53 function and cooperate with each other to regulate p53.
Blocking MDM2-p53 and MDMX-p53 interactions as new cancer therapeutic strategies
Because of their critical and non-redundant role in inhibition of p53, targeting MDM2 and/or MDMX could activate the p53 tumor suppressor function in tumor cells retaining wild-type p53. One attractive approach is to directly block the MDM2-p53 or MDMX-p53 protein-protein interaction using small-molecule inhibitors [40, 41].
The MDM2-p53 interaction was mapped to the first ~120 N-terminal amino acid residues of MDM2 and the first 30 N-terminal residues of p53 [26, 27]. Similarly, the MDMX-p53 interaction also involves the N-terminus of MDMX and the N-terminus of p53. The high-resolution crystal structure of MDM2 in complex with a short p53-peptide (residues 15–29) was published in 1996 (Figure 1A) [42], and the crystal structure of human MDMX bound to the 15-mer peptide derived from human p53 N-terminus domain was reported in 2008 (Figure 1B) [43]. These studies have provided detailed atomic information about the interactions between MDM2-p53 and MDMX-p53. The co-crystal structure of the MDM2-p53 complex clearly reveals that their interaction is mediated by a well-defined hydrophobic surface groove in MDM2 and primarily Phe19, Trp23, and Leu26, three key hydrophobic residues in p53.
As compared to the MDM2-p53 interaction, the MDMX-p53 interaction involves the same three key hydrophobic residues in p53, and a similar but not identical hydrophobic surface groove in MDMX. It is worthy to note that the binding poses of these three key residues of p53 peptide in the two co-crystal structures are very similar (Figure 1C).

In this review, we refer to the three p53 binding pockets on the surface of MDM2 and MDMX as the Phe19, Trp23, and Leu26 pockets. While most residues in the MDM2 and MDMX binding pockets are identical, some residues around the Leu26 pockets are different (Figure 1D), leading to changes in the shape and size of the MDMX groove as compared to the MDM2 groove. These include Met53 (Leu54 in MDM2), Leu98 (Ile99 in MDM2), Leu102 (Ile103 in MDM2), and Pro95 (His96 in MDM2), which result in a shallower and smaller binding groove in MDMX compared to that in MDM2. Additionally, around the Trp23 pocket, the Leu85 residue in MDMX is aligned with the Phe86 residue in MDM2. Overall, the binding groove in MDM2 is compact and well-defined, as is, to a lesser extent, the groove in MDMX. The well-defined binding grooves in MDM2 and MDMX suggest the feasibility of designing non-peptide, small-molecule inhibitors to block the MDM2-p53 interaction and/or the MDMX-p53 interaction, which we refer to as MDM2 inhibitors or MDMX inhibitors in this review.
Several reviews [39-41, 44-50] have discussed progress on the development of MDM2 inhibitors. In this review, we will focus on structure-based strategies for the design of small molecule MDM2 inhibitors and summarize the current progress in the design of MDMX inhibitors.
Structure-based design of MDM2 inhibitors
In 2004, scientists from Hoffmann-La Roche, Inc. reported the first class of potent and specific small molecule MDM2 inhibitors, which they named nutlins (Figure 2) [51].

Nutlin-3a was one of the most potent compounds in this class in the initial report and bound to recombinant MDM2 protein with IC50 = 90 nM in a biochemical binding assay [51]. A co-crystal structure of nutlin-2 complexed with MDM2 (Figure 3A, PDB: 1RV1) showed that the two bromo-substituted phenyl rings of nutlin-2 occupy the Trp23 and Leu26 pockets, and an ethyl substituent on the third phenyl group of nutlin-2 is lodged in the Phe19 pocket. Overlay of nutlin-2 on the p53 peptide showed the two phenyl rings and the ethoxyl group of nutlin-2 nicely superimposed on the three key hydrophobic binding residues of the p53 peptide (Figure 3B).

By occupying the same pockets in MDM2 where p53 binds, MDM2 inhibitors block the MDM2-p53 protein-protein interaction, and hence the MDM2-mediated p53 ubiquitination and degradation of p53, leading to p53 accumulation and transcriptional activation of p53 in cells with wild-type p53 but not in cells with mutated or deleted p53 [51]. Hence, the cellular activity of selective MDM2 inhibitors should be p53-dependent. Indeed, Nutlin-3a effectively inhibited cell growth with IC50 of ~1 µM in p53 wild type cells (SJSA-1, HCT116, and RKO) and had minimal activity at 10 µM in tumor cells harboring mutated p53 (MDA-MB-435 and SW480) [51]. Furthermore, nutlins induced accumulation of p53 in cells with wild-type p53 and transcription of p53-regulated genes, such as p21 and MDM2, but not TP53, in a dose-dependent manner. Significantly, oral administration of nutlin-3 demonstrated 90% inhibition of tumor growth in mice bearing SJSA-1 xenograft tumors with wild-type p53 and amplified MDM2 and mice did not suffer significant weight loss or show other signs of toxicity. The in vitro and in vivo data obtained using nutlins provided the first important proof-of-concept that MDM2 inhibitors may have a therapeutic potential for the treatment of human cancer.
Further structure-based optimization of nutlin-3a has yielded RG7112 (Figure 2), the first MDM2 inhibitor advanced into clinical trials for treating multiple human cancers [52, 53]. There are four key features of the modification of nutlin-3a: (1) Dimethyl groups were introduced into the imidazole ring of nutlins to prevent its oxidation, (2) Isopropyl ether in nutlin-3a was replaced with ethyl ether in RG7112 to reduce molecular weight while retaining the same efficiency for hydrophobic interaction, (3) Methyl phenyl ether, which was found to be the metabolic soft spot, was replaced with tert-butyl group to decrease the metabolic liability, and (4) A polar group, methyl sulfonyl, was added to the urea part to improve MDM2 binding, as well as pharmacokinetic (PK) properties. With these modifications, RG7112 showed enhanced binding affinity to MDM2 with Kd = 10.7 nM and is 3-times more potent than nutlin-3a in inhibition of cell growth. In efficacy experiments, RG7112 at 100 mg/kg daily oral dose achieved partial tumor regression in two human osteosarcomas (SJSA-1 and MHMn) and one human prostate tumor (LNCaP) in mouse xenograft models [53].
Overall, the in vitro and in vivo data obtained from the nutlins demonstrated that potent and specific, non-peptide small-molecule inhibitors can be successfully designed to block the p53-MDM2 interaction and that such inhibitors may have a therapeutic potential for cancer treatment by reactivation of wild-type p53.
Structure-based design of spirooxindole-containing compounds as MDM2 inhibitors
In 2005, the Wang group at the University of Michigan reported the structure-based design of spirooxindole–containing small molecules as a new class of potent small-molecule inhibitors of the MDM2-p53 interaction [54]. In their design, an oxindole group was found to nicely mimic the Trp23 moiety and spirooxindole–containing natural products were identified and docked to the MDM2 protein [54]. Computational docking studies suggested that while these spirooxindole–containing natural products themselves failed to interact effectively with MDM2, the spirooxindole structure could be used as a scaffold for the design of compounds that mimic the three key residues in p53 for binding to MDM2. Compound 1 (MI-5, Figure 4) was designed and predicted to interact effectively with MDM2; the oxindole moiety 1 inserts into the Trp23 pocket, the phenyl group occupies the Phe19 pocket, and the isopropyl group occupies the Leu26 pocket. MI-5 was synthesized using an efficient synthetic procedure and was determined to bind to recombinant MDM2 with Ki = 8.5 µM in a fluorescence polarization (FP) binding assay. Computational modeling further suggested that introduction of a chlorine atom at the meta position in the phenyl group in MI-5 and conversion of the isopropyl group to tert-butyl could enhance the interactions with MDM2. Accordingly, 2 (MI-17) was synthesized and found to bind to MDM2 with Ki = 86 nM, 100-times more potently than the initial compound MI-5. MI-17 was shown to activate p53 in tumor cells with wild-type p53. MI-17 had an IC50 = 830 nM in the LNCaP prostate cancer cell line in a cell growth inhibition assay and was 27-fold less potent against the PC-3 cancer cell line with deleted p53.

Further structure-based optimization of 2 (MI-17) was performed to improve its potency, which resulted in MI-63. MI-63 binds to MDM2 with Ki = 3 nM and has good water solubility [55]. However, MI-63 only has a modest oral bioavailability (10%) in pharmacokinetic studies in rats. Efforts were made to improve the oral bioavailability of MI-63, which resulted in MI-219 [56]. MI-219 binds to MDM2 with Ki = 5 nM and has an oral bioavailability of 65% in rats and a much improved exposure in pharmacokinetic studies in rats. Detailed investigation of MI-219 was performed in vitro and in vivo to determine its therapeutic potential and its molecular mechanism of action [56]. MI-219 was shown to induce activation of p53 by blocking the MDM2-p53 protein-protein interaction in cells expressing wild-type p53 but failed to induce p53 activation in cells expressing mutated p53, consistent with its expected mode of action as a potent and specific MDM2 inhibitor. MI-219 potently inhibited cell growth in cancer cells with wild-type p53 (IC50 values ~ 1 mM) and displayed more than 10-fold selectivity in tumor cells with mutated or deleted p53. A single oral dose of MI-219 induced robust activation of p53 in SJSA-1 xenograft tumor tissue in mice. MI-219 achieved complete tumor growth inhibition in multiple xenograft models of human cancer, although it failed to achieve tumor regression [57]. In normal cells, it induced p53 activation leading to cell cycle arrest but not cell death. MI-219 also activated p53 in normal mouse tissues but caused no tissue damage, even with repeated dosing. The in vitro and in vivo data for MI-219 provided clear evidence that MI-219 is a potent and specific MDM2 inhibitor.
Spirooxindole–containing compounds, such as MI-5, MI-17 and MI-63, have at least four chiral centers in their chemical structures. It was discovered that compound 3, which has a different stereochemistry from MI-17, MI-63, and MI-219, has a very high affinity to MDM2 (Ki < 1 nM). Further optimization of the pharmacokinetic properties of 3 yielded MI-888, which has a Ki value of 0.44 nM and is highly potent and selective in inhibition of tumor cell growth in cancer cell lines with wild-type p53 over those with mutated or deleted p53 [58]. MI-888 has an excellent oral bioavailability in rats and is capable of achieving complete and durable tumor regression in two animal models of human cancer upon daily oral administration. An analogue of MI-888 has been advanced into Phase I clinical development.
Structure-based design of piperidinone-containing MDM2 inhibitors by Amgen
Recently, scientists from Amgen have reported the structure-based design of a new class of potent MDM2 inhibitors [59]. Based upon several classes of known MDM2 inhibitors, including nutlins and spirooxindole–containing compounds, compound 4 was designed featuring a 1,3,5,6-tetrasubstituted piperidinone scaffold (Figure 5).
![Figure 5. Representative examples of piperidinone compounds from Amgen (see reference [59] for HTRF-based binding assay conditions). Figure 5](https://web.archive.org/web/20160322125634im_/http://biodiscoveryjournal.co.uk/Archive/Media/A30Figure-5_550.jpg)
While 4 had an IC50 value of 2420 nM in an MDM2 binding assay, compound 5, a diastereoisomer of 4, had an IC50 = 34 nM, >50-times more potent than 4. Computational docking showed that the p-chloro substituted phenyl ring occupied the Trp23 pocket, m-chloro substituted phenyl ring was in the Leu26 pocket, and the cyclopropyl group occupied the Phe19 pocket. The carboxylic acid group at the 3-position of piperidinone, was found to have additional interaction with the His96 residue of MDM2. Deletion of the carboxylic acid group led to a 44-fold loss in binding affinity, emphasizing the significance of this group. Replacement of the cyclopropyl group in 5 by a chiral tert-butyl 2-butanoate, yielded 6, which is 8-times more potent than 5 to MDM2. A co-crystal structure of 6 in a complex with MDM2 (Figure 6, PDB: 4ERE) confirmed the predictions from the computational docking.

Consistent with the initial computational docking prediction, the two phenyl groups inserted into the Leu26 and Trp23 pockets, while the ethyl side moiety of the tert-butyl 2-butanoate group was directed towards the Phe19 pocket, and the carboxylic acid formed a salt bridge with His96. The crystal structure also showed that the two phenyl groups had a gauche-like orientation, which was less favored over the more stable anti-like conformation as suggested by quantum mechanical calculations. The same quantum mechanical calculations also suggested that installation of two substituents at the C3 position of the piperidinone ring would favor the binding conformation (gauche-like) over non-binding confirmation (anti-like) by 3:1. Guided by these insights, 7 was designed and synthesized by adding an extra methyl group at C3 of the piperidinone ring and found to be about twice as potent as 6.
In addition to all the features mentioned above, this class of MDM2 inhibitors, such as 5, induced ordering of N-terminal residues of MDM2, as shown by NMR and X-ray crystallography studies (PDB: 2LZG and 4HBM, respectively). The 14−24 residues of MDM2 adopted α-helix (21-24), β-turn (17-20), and β-sheet (14-16) structures upon binding to the piperidinones, whereas these N-terminal residues were un-structured in apo-MDM2. The back-folding of the β-sheet gained additional van der Waals interaction between meta-Cl phenyl moiety of piperidinones and Val14 and Thr16 [60]. Compound 7 indeed binds to MDM2 with a very high affinity with IC50 = 2.2 nM. Further modification of the tert-butyl ester of 7 into a hydroxyl group yielded MI-8553, which had an IC50 = 1.1 nM to MDM2. In a mouse SJSA-1 tumor xenograft model, oral gavage of AM-8553 at 200 mg/kg once daily achieved partial tumor regression, demonstrating its strong antitumor activity.
1,4-Diazepines as MDM2 inhibitors
In 2005, a class of benzodiazepine compounds was reported by Johnson & Johnson as MDM2 inhibitors [61, 62]. The initial hit compounds, N,N-dibenzylcinnamoyl amide 8 and N,N-dibenzylbenzamide 10 (Figure 7), were identified through a high-throughput screening assay [63].

Compounds 8 and 10 bound to MDM2 with IC50 = 30 µM and 15 µM, respectively. Two cyclization strategies were pursued to optimize these initial hits [64, 65]. Cyclization of 10 yielded 1,4-benzodiazepine-2,5-dione (BZD) 11, which binds to MDM2 with IC50 = 1.7 µM. Optimization of the substituents on the three phenyl rings resulted in racemic mixture of 12 and 13, which had an MDM2 binding IC50 = 0.42 µM. While the R,R-isomer, 13, has Kd = 4.8 µM, the S,S-isomer 12 has Kd = 80 nM and is 50-times more potent than 13. The stereogenic center at the α-carbonyl position of 12 was liable to epimerize under basic conditions. Consequently, 14 and 15 were made to address the epimerization issue. Compound 14 was as potent as the racemic mixture of 12 and 13, but compound 15 was inactive.
The crystal structure of the complex formed by MDM2 and BZD 12 (S,S-isomer) was obtained (Figure 8, PBD: 1T4E) at a resolution of 2.7 Å, [64] which showed that 12 closely mimics the three key p53 binding residues for interaction with MDM2. The iodobenzene ring occupied the Phe19 pocket, the p-chlorophenyl occupied the Trp23 pocket, and the other p-chlorophenyl, which was at the α-position relative to the carboxylic acid group, occupied the Leu26 pocket.

In cells, 12 induced clear dose-dependent accumulation of p53 and its targeted gene products, p21 and MDM2, at concentrations of 5-10 µM in JAR cells with wild-type p53. Compound 12 decreased cell proliferation with IC50 = 30 µM in JAR cells and did not inhibit proliferation of MDA-MB-231 cells with mutated p53, demonstrating the specificity.
Pharmacokinetic studies showed that 12 had rapid in vivo clearance and was not orally bioavailable [65]. Calculations indicated only 0.002% of neutral species of 12 at physiological pH (7.4), largely because of the presence of the carboxylic acid group and subsequent modifications aimed at improving in vitro potency and cellular activity focused on the carboxylic acid site. SAR showed that extension of the carboxylic acid group or alkylation of the amide nitrogen did not improve potency in mammary carcinoma MCF7 cells with wild-type p53. Replacement of the carboxylic acid with a methyl group followed by alkylation of the NH group with a 4-carboxybutyl group furnished compound 16, which had MDM2 binding affinity IC50 = 700 nM and had IC50 = 7 µM in MCF7 cells with wild-type p53 in a BrdU incorporation assay. Oral administration of 16 in mice at 40 mg/kg gave a Cmax value of 6.3 µM and a Tmax value of 6 h. 16 had a plasma clearance t1/2 of 2.5 h and 100% oral bioavailability.
Additional modification and analysis suggested that the addition of an NH2 group to the o-position of one of the phenyl rings could possibly gain an additional hydrogen bond with Val93 of MDM2 [66, 67]. In addition, the 4-carboxybutyl group could be replaced with a 2-(2-methoxyethoxy) ethyl group without significantly attenuating the cell potency [66]. Finally, an asymmetric synthesis of 1,4-benzodiazepine-2,5-dione compounds was developed and optically pure BZD 17 was prepared [67]. Compound 17 was the best of this class, with an MDM2 binding IC50 = 394 nM. It had IC50 = 1.1 µM in the MCF-7 cell line in a BrdU incorporation assay and was 50-fold more selective over the MDA-MB-213 cell line with mutated p53. Despite these significant efforts, this class of MDM2 inhibitors did not show strong antitumor activity in animal models of human cancer and did not progress into clinical development.
Structure-based design of isoindolinones as MDM2 inhibitors
In 2005, researchers from University of Newcastle reported the design of MDM2 inhibitors based on the isoindolinone scaffold [68]. Guided by computational docking studies, extensive modifications of isoindolinones 18-20 were performed (Figure 9) [69].
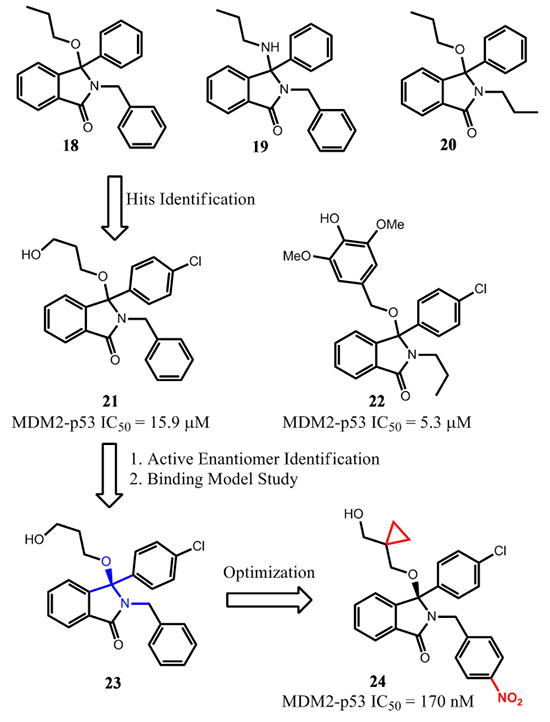
These efforts led to the identification of compounds 21 and 22, which bind to MDM2 with IC50 = 15.9 and 5.3 µM, respectively. Treatment of SJSA cells with either compound activated p53 at 5-10 µM as measured by Western blot analysis. In addition, dose-dependent activation of p53 was achieved by treatment of 21 in a range of 5-40 µM in a luciferase-based, p53-dependent reporter gene assay. Subsequently, a structure-based optimization was performed to improve the binding affinity to MDM2 and cellular potency for this class of compounds [70].
Facilitated by NMR analysis of the ligand-protein interaction in solution, the Leu54 region of MDM2 was found to have relatively stronger interaction with the 3-hydroxypropoxy moiety of 23, a chiral version of 21, which became the focus of further modifications on the 3-hydroxypropoxy group. Compound 24, the (+)-R-enantiomer, was one of the best compounds and had an IC50 of 0.17 µM in an ELISA binding assay. Interestingly, unlike the p53 peptide and other MDM2 inhibitors, the binding model of 24 was unique that it only occupied two hydrophobic binding pockets; the 4-chlorophenyl group occupied the Leu26 pocket, and the isoindolinone scaffold occupied the Trp23 pocket. The other two functional groups of 24 had polar interactions with MDM2. The nitrophenyl group was close to the Gln59 residue and the free hydroxyl group, as designed, projected towards the Leu54 pocket. Nevertheless, upon treatment of 24 at 1-10 µM for 4 h, robust activation of p53 and its targeted gene products, p21 and MDM2, were observed in SJSA-1 cells. The enantiomer of 24 was inactive in the same assay at concentrations as high as 20 µM. The cell growth inhibition GI50 of 24 in p53 isogenic cell lines, HCT116 p53+/+ and HCT116 p53-/-, were close (20 µM and 24 µM, respectively), suggesting that the cytotoxicity for compound 24 may be a combination of both on- and off-target effects. Further optimization of this class of compounds will be needed to obtain compounds suitable for in vivo studies and for potential clinical development.
Other small molecule MDM2 inhibitors based on nutlins
Recently, two classes of small molecule MDM2 inhibitors were generated from modifications of nutlins. The first, reported by Hu et. al. replaced the amide side chain of nutlin-3a with a carbamate group (Figure 10) [71]. One of the most potent compounds (25) showed MDM2 binding affinity and cellular activity similar to those of nutlin-3a.

The second class was reported in 2012 by scientists from the Daiichi Sankyo [72, 73]. The key modification in this case was the replacement of the imidazoline core of the nutlins by a fused bicyclic dihydroimidazothiazole, a modification which decreased the MDM2 binding affinity of 26 to IC50 = 260 nM, vs nutlin-3a IC50 = 90 nM. Interestingly, installation of one methyl group on the imidazoline core improved MDM2 binding affinity of 27 (IC50 = 92 nM) and suppressed oxidation of the partially saturated 5,6-dihydroimidazo[2,1-b]thiazole to unsaturated imidazo[2,1-b]thiazole. In addition, replacement of the piperazin-2-one moiety with a proline ring yielded compound 28 that had a binding affinity IC50 = 9.2 nM to MDM2, 10-times more potent than nutlin-3a.
A co-crystal structure of MDM2-28 (Figure 11, PDB: 3VZV) showed that two para-chlorophenyl groups occupied the Leu26 and Trp23 pockets and the isopropyl group occupied the Phe19 pocket. Overlaying the co-crystal structure of 28-MDM2 complex on that of nutlin-MDM2 showed one critical difference: the proline moiety of 28 induced conformational changes in the MDM2 loop in the region from Tyr67 to Gln72, providing additional hydrophobic interactions between the proline and MDM2, which may contribute to the enhanced MDM2 binding affinity of 28 as compared to nutlins. Although no detailed biological data are reported to date, these new nutlin analogues represent potent and potentially promising MDM2 inhibitors.

The structural basis for rational design of MDMX inhibitors
In 2010, Popowicz and coworkers reported the first small molecule-human MDMX co-crystal structure at a resolution of 1.50 Å (Figure 12A, PDB: 3LBJ) [74]. This provided an opportunity for the first time to study details at the atomic level of a small molecule bound to MDMX.

The small molecule, WK298 (Figure 13), had a moderate MDMX binding affinity (Ki = 11 µM) and a higher affinity for MDM2 (Ki = 109 nM). The three p53 binding pockets Phe19, Trp23 and Leu26 were occupied by the phenyl, 6-chloroindole, and 4-chlorobenzyl groups of WK298, respectively. Two additional hydrogen bond interactions were found between WK298 and Met53 and His54. The aliphatic side chain of WK298 also had a hydrophobic interaction around the binding cleft close to the Gly57-Met61 region.

A co-crystal structure of MDM2 and WK23 (Figure 12B, PDB: 3LBK), an analogue of WK298, was also determined at a resolution of 2.3 Å and this facilitated analysis of the differences between MDM2-p53 and MDMX-p53. WK298 and WK23 had nearly identical poses as revealed in their crystal structures, but the MDM2 and MDMX proteins had significant differences around the Leu26 binding site, corresponding to the His96-Tyr100 region of MDM2 and the Pro95-Tyr99 region of MDMX. In MDM2, the α-helix bundle of the His96-Tyr100 region folded more tightly to the Leu26 pocket than the Pro95-Tyr99 region in MDMX. As a result, the Leu26 pocket is less open to solvent in the case of MDM2 and this may explain why WK23 and WK298 bind to MDM2 with much higher affinities than to MDMX.
Other small molecule inhibitors of the MDMX-p53 interaction
Several research groups have reported efforts to discover potent MDMX small molecule inhibitors [71, 75-78]. For example, Reed and co-workers used high-throughput screening to assess a library of 295,848 unique compounds using an in vitro fluorescence polarization binding assay [77]. The best compound reported, SJ-172550 (Figure 14), had an MDMX binding affinity IC50 = 0.84 µM, while the affinity of nutlin-3a was 20.1 µM in the same assay. Further investigation revealed that SJ-172550 formed a reversible covalent complex with MDMX [79].
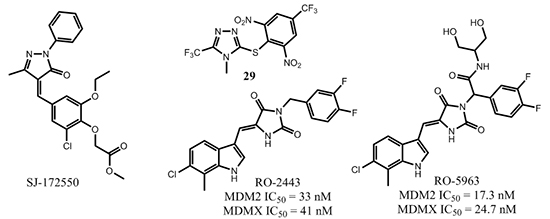
Tanaka and co-workers screened a library of 40,000 compounds to identify MDMX inhibitors [78]. One of the compounds identified, 29, had an MDMX binding affinity IC50 of 0.5 µM in the surface plasmon resonance (SPR) assay and 1.2 µM in fluorescence correlation spectroscopy (FCS) assay while nutlin-3 had an IC50 of 50 µM in the SPR assay and 20 µM in the FCS assay to MDMX, respectively. Compound 29 had a cell growth inhibitory GI50 of 1.6 µM in the p53 wild-type MV4;11 leukemia cell line, 5.5 µM in the p53-deficient HI299 cell line, and 9.8 µM in WI-38 cultured normal human cells, respectively, thus only displaying modest selectivity for cancer cells with different p53 status.
Recently, Vassilev and coworkers reported a dual small molecule (RO-2443) that bound to MDMX with an IC50 of 41 nM and to MDM2 with an IC50 of 33 nM, while nutlin-3a bound to MDMX with an IC50 of 9.3 µM [80]. A crystal structure of the MDMX-RO-2443 complex (PBD: 3U15) was obtained at 1.8 Å resolution and showed that two MDMX protein molecules formed a dimer. Two of the p53-peptide binding pockets formed a hollow core and within this core, there were two molecules of RO-2443 parallel to each other in a head-to-tail fashion. Overlay of the crystal structure on the p53 peptide showed that a difluorophenyl group of one RO-2443 occupied the Trp23 binding pocket and a chloroindole group of the other RO-2443 occupied the Phe19 pocket, leaving the Leu26 pocket vacant. A related compound, RO-5963, developed as a water-soluble version of RO-2443 also had high binding affinities to MDM2 and MDMX, with IC50 values of 17.3 nM and 24.7 nM, respectively. The activity of RO-5963 was p53-dependent. RO-5963 was active in p53 wild-type cell lines (IC50 = 2-3 µM in MCF-7, HCT-116, and RKO) but inactive in p53 mutant cell lines (IC50 > 10 µM in SW480 and MDA-MB435). In MDMX-overexpressing breast cancer MCF7 cells, RO-5963 showed cell growth inhibition with IC50 = 2 µM, stabilized p53 in a dose-dependent manner, and elevated protein levels of its transcription targets, p21 and MDM2. The binding mode of RO compounds is interesting because although the Leu26 pocket was not occupied, these compounds had high affinity to both MDM2 and MDMX proteins and showed strong activity in p53 wild-type cancer cell lines. Further optimization of RO-5963 could potentially yield highly potent and efficacious small molecule MDMX/MDM2 dual inhibitors.
Clinical development of small molecule MDM2 inhibitors
The first MDM2 inhibitor advanced into clinical development was RG-7112 [53] (Figure 2), a nutlin. The clinical trials of RG-7112 (RO5045337) covered a wide range of conditions such as sarcoma, myelogenous leukemia, neoplasms, and hematologic neoplasms. (ClinicalTrials.gov Identifiers: NCT00559533, NCT00623870, NCT01677780, NCT01164033, NCT01605526, NCT01143740, and NCT01635296). The results from phase I trials in patients with MDM2-amplified liposarcoma have been recently reported [81]. While showing only limited tumor volume reduction, there was clear evidence of p53 activation and cell growth inhibition in MDM2-amplified liposarcomas. Hoffman-La Roche has also advanced a second small molecule MDM2 inhibitor (RO5503781) into phase I clinical trials, in which the drug is being tested as a single agent or in combination with cytarabine (ClinicalTrials.gov Identifier: NCT01773408 and NCT01462175).
In addition to these MDM2 inhibitors from Roche, there are four other compounds now in clinical development. MK-8242, also known as SCH 900242 [82], is being tested as a single agent in patients with advanced solid tumors or acute myelogenous leukemia in two phase I clinical trials (ClinicalTrials.gov Identifier: NCT01463696 and NCT01451437) and in combination with cytarabine in those with acute myelogenous leukemia. SAR405838, analogue of MI-888, discovered in the Wang laboratory at the University of Michigan, has been advanced into phase I clinical trials by Sanofi S.A in 2012 (ClinicalTrials.gov Identifier: NCT01636479). CGM097, developed by Novartis, has also progressed into phase I clinical trials in 2013 (ClinicalTrials.gov Identifier: NCT01760525). Very recently, Daiichi Sankyo Inc. has initiated phase I clinical trial of DS-3032b (ClinicalTrials.gov Identifier: NCT01877382).
Summary and outlook
Although targeting the MDM2-p53 protein-protein interaction by non-peptide, small-molecule inhibitors has proven to be considerably more difficult than targeting receptors and enzymes, major advancement has been made in the last decade. The design of potent, non-peptide small-molecule inhibitors of the MDM2-p53 interaction has been assisted greatly by the determination of high-resolution co-crystal structures of MDM2 complexed with p53 peptides and several classes of non-peptide small-molecule inhibitors, and by effective use of computational, structure-based design tools. In general, the development of inhibitors has focused on three critical p53 residues, and most small molecules successfully occupy the binding site for all three residues (Figure 15).

At present, non-peptide, small-molecule inhibitors with very high affinities to MDM2 (Ki < 1 nM) and optimal pharmacokinetic properties have been developed. In animal models of human cancer, potent antitumor activity, including complete tumor regression, has been observed for some of these MDM2 inhibitors with high affinity to MDM2 and optimized pharmacokinetics. To date, six such compounds have progressed into clinical development. p53 activation was achieved by the first clinical MDM2 inhibitor RG-7112. Both single agents and combinations with other agents are being tested in clinical trials for these MDM2 inhibitors.
In addition to MDM2, MDMX is another negative regulator of p53 activity. Encouraged by the success in the design of non-peptide, small-molecule inhibitors of the MDM2-p53 interaction, there are considerable efforts in the design and development of non-peptide, small-molecule inhibitors of the MDMX-p53 interaction. Compounds with binding affinities to MDMX in the nanomolar range have been recently reported, suggesting the possibility of designing highly potent MDMX inhibitors for potential therapeutic applications.
Acknowledgements
We are grateful for financial support from the National Cancer Institute, National Institutes of Health (Grants R01CA121279, P50CA06956, and P50CA097248), the University of Michigan Cancer Center (Core Grant P30CA046592), Ascenta Therapeutics, Inc. and Sanofi S.A.
References
- Teodoro JG, Evans SK & Green MR. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J Mol Med (Berl) 2007; 85: 1175-1186.
REFERENCE LINK - Fridman J S & Lowe SW. Control of apoptosis by p53. Oncogene 2003; 22: 9030-9040.
REFERENCE LINK - Vousden KH & Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer 2002; 2: 594-604.
REFERENCE LINK - Lane DP & Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979; 278: 261-263.
REFERENCE LINK - DeLeo AB, Jay G, Appella E, Dubois GC, Law LW & Old LJ. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci U S A 1979; 76: 2420-2424.
REFERENCE LINK - Linzer DI & Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979; 17: 43-52.
REFERENCE LINK - Oren M & Levine AJ. Molecular cloning of a cDNA specific for the murine p53 cellular tumor antigen. Proc Natl Acad Sci U S A 1983; 80: 56-59.
REFERENCE LINK - Feki A & Irminger-Finger I. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol 2004; 52: 103-116.
REFERENCE LINK - Momand J, Zambetti GP, Olson DC, George D & Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992; 69: 1237-1245.
REFERENCE LINK - Fakharzadeh SS, Trusko SP & George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 1991; 10: 1565-1569.
- Fakharzadeh SS, Rosenblum-Vos L, Murphy M, Hoffman EK & George DL. Structure and organization of amplified DNA on double minutes containing the mdm2 oncogene. Genomics 1993; 15: 283-290.
REFERENCE LINK - Hainaut P & Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 2000; 77: 81-137.
REFERENCE LINK - Freedman DA, Wu L & Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci 1999; 55: 96-107.
REFERENCE LINK - Wu X, Bayle JH, Olson D & Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7: 1126-1132.
REFERENCE LINK - Juven-Gershon T & Oren M. Mdm2: the ups and downs. Mol Med 1999; 5: 71-83.
- Jones SN, Roe AE, Donehower LA & Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206-208.
REFERENCE LINK - Montes de Oca Luna R, Wagner DS & Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203-206.
REFERENCE LINK - Ganguli G, Abecassis J & Wasylyk B. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J 2000; 19: 5135-5147.
REFERENCE LINK - Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591-602.
REFERENCE LINK - Oliner JD, Kinzler KW, Meltzer PS, George DL & Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992; 358: 80-83.
REFERENCE LINK - Zhou M, Gu L, Abshire TC, Homans A, Billett AL, Yeager AM & Findley HW. Incidence and prognostic significance of MDM2 oncoprotein overexpression in relapsed childhood acute lymphoblastic leukemia. Leukemia 2000; 14: 61-67.
REFERENCE LINK - Rayburn, E., Zhang, R., He, J. & Wang, H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets 2005; 5: 27-41.
REFERENCE LINK - Momand J, Jung D, Wilczynski S & Niland J. The MDM2 gene amplification database. Nucleic Acids Res 1998; 26: 3453-3459.
REFERENCE LINK - Gunther T, Schneider-Stock R, Hackel C, Kasper HU, Pross M, Hackelsberger A et al. Mdm2 gene amplification in gastric cancer correlation with expression of Mdm2 protein and p53 alterations. Mod Pathol 2000; 13: 621-626.
REFERENCE LINK - Bond GL, Hu W & Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets 2005; 5: 3-8.
REFERENCE LINK - Capoulade C, Bressac-de Paillerets B, Lefrere I, Ronsin M, Feunteun J, Tursz T et al. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene 1998; 16: 1603-1610.
REFERENCE LINK - Momand J, Wu HH & Dasgupta G. MDM2–master regulator of the p53 tumor suppressor protein. Gene 2000; 242: 15-29.
REFERENCE LINK - Kemp CJ, Donehower LA, Bradley A & Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 1993; 74: 813-822.
REFERENCE LINK - Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 1997; 43: 34-42.
REFERENCE LINK - Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996; 15: 5349-5357.
- Marine JC & Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun 2005; 331: 750-760.
REFERENCE LINK - Stad R, Ramos YF, Little N, Grivell S, Attema J, van Der Eb AJ et al. Hdmx stabilizes Mdm2 and p53. J Biol Chem 2000; 275: 28039-28044.
- Migliorini D, Danovi D, Colombo E, Carbone R, Pelicci PG & Marine JC. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J Biol Chem 2002; 277: 7318-7323.
REFERENCE LINK - Linke K, Mace PD, Smith CA, Vaux DL, Silke J & Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008; 15: 841-848.
REFERENCE LINK - Wade M & Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res 2009; 7: 1-11.
REFERENCE LINK - Wang X, Wang J & Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 2011; 286: 23725-23734.
REFERENCE LINK - Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A 2006; 103: 3232-3237.
REFERENCE LINK - Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A 2011; 108: 12001-12006.
REFERENCE LINK - Riedinger C & McDonnell JM. Inhibitors of MDM2 and MDMX: a structural perspective. Future Med Chem 2009; 1: 1075-1094.
REFERENCE LINK - Brown CJ, Lain S, Verma CS, Fersht AR & Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009; 9: 862-873.
REFERENCE LINK - Wade M, Li YC & Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83-96.
REFERENCE LINK - Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996; 274: 948-953.
REFERENCE LINK - Popowicz GM, Czarna A & Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle 2008; 7: 2441-2443.
- Lane DP, Cheok CF & Lain S. p53-based Cancer Therapy. Cold Spring Harbor Perspectives in Biology 2010; 2: 1-24.
REFERENCE LINK - Millard M, Pathania D, Grande F, Xu S & Neamati N. Small-molecule inhibitors of p53-MDM2 interaction: the 2006-2010 update. Curr Pharm Des 2011; 17: 536-559.
REFERENCE LINK - Popowicz GM, Domling A & Holak TA. The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew Chem Int Ed 2011; 50: 2680-2688.
REFERENCE LINK - Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med 2007; 13: 23-31.
REFERENCE LINK - Khoury K, Popowicz GM, Holak TA & Domling A. The p53-MDM2/MDMX axis – A chemotype perspective. MedChemComm 2011; 2: 246-260.
REFERENCE LINK - Dickens MP, Fitzgerald R & Fischer PM. (2010). Small-molecule inhibitors of MDM2 as new anticancer therapeutics. Sem Cancer Biol 2010; 20: 10-18.
REFERENCE LINK - Wang S, Zhao Y, Bernard D, Aguilar A & Kumar S. Targeting the MDM2-p53 Protein-Protein Interaction for New Cancer Therapeutics. Top Med Chem 2012; 8: 57-80.
REFERENCE LINK - Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844-848.
REFERENCE LINK - Vu B, Wovkulich P, Pizzolato G, Lovey A, Ding Q, Jiang N, Liu J-J et al. Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development. ACS Med Chem Lett 2013; 4: 466-469.
REFERENCE LINK - Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M. et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res 2013; 73: 2587-2597.
REFERENCE LINK - Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding YS, Gao W et al. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc 2005; 127: 10130-10131.
REFERENCE LINK - Ding K, Lu YP, Nikolovska-Coleska Z, Wang GP, Qiu S, Shangary S et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem 2006; 49: 3432-3435.
REFERENCE LINK - Yu SH, Qin DG, Shangary S, Chen JY, Wang GP, Ding K et al. Potent and Orally Active Small-Molecule Inhibitors of the MDM2-p53 Interaction. J Med Chem 2009; 52: 7970-7973.
REFERENCE LINK - Shangary S, Qin D, McEachern D, Liu M, Miller R. S, Qiu S et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A 2008; 105: 3933-3938.
- Zhao Y, Yu S, Sun W, Liu L, Lu J, McEachern D et al. A Potent Small-Molecule Inhibitor of the MDM2-p53 Interaction (MI-888) Achieved Complete and Durable Tumor Regression in Mice. J Med Chem 2013; 56: DOI: 10.1021/jm4005708.
REFERENCE LINK - Rew Y, Sun DQ, De Turiso FGL, Bartberger MD, Beck HP, Canon J et al. Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem 2012; 55: 4936-4954.
REFERENCE LINK - Michelsen K, Jordan JB, Lewis J, Long AM, Yang E, Rew Y et al. Ordering of the N-terminus of human MDM2 by small molecule inhibitors. J Am Chem Soc 2012; 134: 17059-17067.
REFERENCE LINK - Parks DJ, Lafrance LV, Calvo RR, Milkiewicz KL, Gupta V, Lattanze J et al. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction: discovery and SAR. Bioorg Med Chem Lett 2005; 15: 765-70.
REFERENCE LINK - Raboisson P, Marugan JJ, Schubert C, Koblish HK, Lu T, Zhao S et al. Structure-based design, synthesis, and biological evaluation of novel 1,4-diazepines as HDM2 antagonists. Bioorg Med Chem Lett 2005; 15: 1857-1861.
REFERENCE LINK - Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. Journal of Biomolecular Screening 2001; 6: 429-440.
REFERENCE LINK - Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem 2005; 48: 909-912.
REFERENCE LINK - Parks DJ, LaFrance LV, Calvo RR, Milkiewicz KL, Marugan JJ, Raboisson P et al. Enhanced pharmacokinetic properties of 1,4-benzodiazepine-2,5-dione antagonists of the HDM2-p53 protein-protein interaction through structure-based drug design. Bioorg Med Chem Lett 2006; 16: 3310-3314.
REFERENCE LINK - Leonard K, Marugan JJ, Raboisson P, Calvo R, Gushue JM, Koblish HK et al. Novel 1,4-benzodiazepine-2,5-diones as Hdm2 antagonists with improved cellular activity. Bioorg Med Chem Lett 2006; 16: 3463-3468.
REFERENCE LINK - Marugan JJ, Leonard K, Raboisson P, Gushue JM, Calvo R, Koblish HK et al. Enantiomerically pure 1,4-benzodiazepine-2,5-diones as Hdm2 antagonists. Bioorg Med Chem Lett 2006; 16: 3115-3120.
REFERENCE LINK - Hardcastle IR, Ahmed SU, Atkins H, Calvert AH, Curtin NJ, Farnie G et al. Isoindolinone-based inhibitors of the MDM2-p53 protein-protein interaction. Bioorg Med Chem Lett 2005; 15: 1515-1520.
REFERENCE LINK - Hardcastle IR, Ahmed SU, Atkins H, Farnie G, Golding BT, Griffin RJ et al. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction based on an isoindolinone scaffold. J Med Chem 2006; 49: 6209-6221.
REFERENCE LINK - Hardcastle IR, Liu JF, Valeur E, Watson A, Ahmed SU, Blackburn TJ et al. Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J Med Chem 2011; 54: 1233-1243.
REFERENCE LINK - Wang H, Ma X, Ren S, Buolamwini JK & Yan C. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 2011; 10: 69-79.
REFERENCE LINK - Miyazaki M, Naito H, Sugimoto Y, Kawato H, Okayama T, Shimizu H et al. Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold. Bioorg Med Chem Lett 2013; 23: 728-732.
REFERENCE LINK - Miyazaki M, Kawato H, Naito H, Ikeda M, Miyazaki M, Kitagawa M et al. Discovery of novel dihydroimidazothiazole derivatives as p53-MDM2 protein-protein interaction inhibitors: synthesis, biological evaluation and structure-activity relationships. Bioorg Med Chem Lett 2012; 22: 6338-6342.
REFERENCE LINK - Popowicz GM, Czarna A, Wolf S, Wang K, Wang W, Domling A et al. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle 2010; 9: 1104-1111.
REFERENCE LINK - Bold G, Furet P, Gessier F, Kallen J, Hergovich Lisztwan J, Masuya K et al. Tetra-substituted heteroaryl compounds and their use as MDM2 and/or MDM4 modulators. PCT Patent WO/2011/023677.
- Uoto K, Kawato H, Sugimoto Y, Naito H, Miyazaki M, Taniguchi T et al.. Imidazothiazole derivative having 4,7-diazaspiro[2.5]octane ring structure. PCT Patent WO/2009/151069.
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 2010; 285: 10786-10796.
REFERENCE LINK - Tsuganezawa K, Nakagawa Y, Kato M, Taruya S, Takahashi F, Endoh M. et al. A Fluorescent-Based High-Throughput Screening Assay for Small Molecules That Inhibit the Interaction of MdmX with p53. Journal of Biomolecular Screening 2013; 18: 191-198.
REFERENCE LINK - Bista M, Smithson D, Pecak A, Salinas G, Pustelny K, Min J et al. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. PLoS One 2012; 7: e37518.
REFERENCE LINK - Graves B, Thompson T, Xia MX, Janson C, Lukacs C, Deo D et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci U S A 2012; 109: 11788-11793.
REFERENCE LINK - Ray-Coquard I, Blay J Y, Italiano A, Le Cesne A, Penel N, Zhi J et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 2012; 13: 1133-1140.
- Perez-Moreno P, Brambilla E, Thomas R & Soria JC. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res 2012; 18: 2443-2451.
REFERENCE LINK



