Abstract
The ability of a cell to conserve and maintain its native DNA sequence is fundamental for the survival and normal functioning of the whole organism and protection from cancer development. Here we review recently obtained results and current topics concerning the role of the ataxia-telangiectasia mutated (ATM) protein kinase as a damage sensor and its potential as therapeutic target for treating cancer. This monograph discusses DNA repair mechanisms activated after DNA double-strand breaks (DSBs), i.e. non-homologous end joining, homologous recombination and single strand annealing and the role of ATM in the above types of repair. In addition to DNA repair, ATM participates in a diverse set of physiological processes involving metabolic regulation, oxidative stress, transcriptional modulation, protein degradation and cell proliferation. Full understanding of the complexity of ATM functions and the design of therapeutics that modulate its activity to combat diseases such as cancer necessitates parallel theoretical and experimental efforts. This could be best addressed by employing a systems biology approach, involving mathematical modelling of cell signalling pathways.
Citation: Khalil HS, Tummala H, Zhelev N. ATM in focus: A damage sensor and cancer target. Biodiscovery 2012; 5: 1.; DOI: 10.7750/BioDiscovery.2012.5.1
Copyright: © 2012 Khalil et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, provided the original authors and source are credited.
Received: September 3, 2012; Accepted: November 28, 2012; Available online /Published: November 30, 2012
*Corresponding Author: Nikolai Zhelev – E-mail: n.zhelev@abertay.ac.uk
1. Introduction
1.1. Mechanisms of Eukaryotic DNA repair
Conservation and protection of the native DNA sequence and structure is essential for the survival and normal functioning of a cell and ultimately of the whole organism. However, our bodies are constantly exposed to diverse types of genotoxic insults from various sources that can damage cellular DNA and threaten the survival. The causes and sources for DNA damage can be both exogenous and endogenous. Major exogenous sources include exposure to ionizing radiation e.g. UV, X-rays, gamma rays [1, 2] and through contact with certain chemical carcinogens. Endogenous means of DNA damage may include generation of reactive oxygen species resulting from metabolic by-products during cellular respiration, mechanical damage to chromosomes e.g. when dicentric or catenated chromosomes are pulled to opposite poles during mitosis, during programmed genomic rearrangements induced by nucleases and defective metabolism of chromosomal ends [3]. The different sources of DNA damage can cause a wide variety of DNA lesions. For example, ionizing radiation can induce single or double stranded DNA breaks (SSBs or DSBs respectively), UV light can cause the formation of pyrimidine dimers or depurination, chemical carcinogens can cause DNA cross links, enzyme-mediated base removal may produce abasic sites, and spontaneous cytosine deamination can result in non-native DNA base, uracil. Additionally, replication fork arrest caused by SSBs can result in its collapse and creation of DSBs [4]. In most of the instances, generation of DNA lesions causes structural and functional changes in DNA that can accumulate through cell division and in extreme cases, leads to cancer and death. Generation of DSBs is generally regarded as the most toxic of all DNA lesions while its repair, as the most complex process [5]. Errors in the repair of DSBs can result in deletion or insertion mutations, chromosomal translocation, and genomic instability leading to malignancy [6].
In order to maintain genomic integrity, eukaryotic cells have developed intricate DNA damage sensing and repair mechanism that combats the diverse sources of DNA damage and ensures survival. Critical features of this highly robust repair mechanism are the ability to specifically recognize the DNA lesion and efficiently remove it. Additionally, the different repair responses that are generated and the resulting network signalling triggered within the cell not only repairs the DNA lesion, but is also tightly linked with the cellular machinery that governs cell-fate decision e.g. cell cycle arrest to promote survival or programmed cell death. DNA repair capacity and response depends on the differentiation state of the cell and the scale and type of damage [7-9]. Depending upon the scale and type of DNA damage, different repair responses can be activated with different outcomes for the cell. Figure 1 shows a simplified illustration of different components of the repair pathway and its effects on cell-fate.
![Figure 1. Cellular responses to DNA damage. Depending upon the scale of DNA damage, one of the two pathways may be activated. If the DNA damage is minor, sensor proteins are activated, which recruit specialized DNA repair enzymes to the damaged site and DNA is repaired. However, if the DNA damage is extensive, the DNA Damage Response (DDR) pathway is activated , which triggers specialized transducer proteins that amplify the damaged signal and activate effector proteins. The effector proteins cause cell cycle arrest and increase in the production of DNA damage repair enzymes. The arrest at the cell cycle checkpoints may result in temporary halting of the cell cycle, a permanent arrest or induction of apoptotic pathways. Source - Khalil HS et al. [10]. Figure 1](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-1_550.jpg)
There are six damage-type-specific repair mechanisms that can be triggered for dealing with different kinds of DNA damage. Table 1 shows the different types of DNA damage and the type-specific DNA repair mechanism that is activated as a result of it.

Since this monograph focuses on regulation and functional elucidation of ATM, a protein kinase involved in DSB repair, DNA repair mechanisms activated only after DSBs, i.e. non-homologous end joining, homologous recombination and single strand annealing will be discussed in more detail.
1.2. Double stranded DNA repair
1.2.1. Non-homologous end joining (NHEJ) DNA repair
This mode of DNA repair is often described as error-prone. Double stranded breaks produced by nucleases or after exposure to ionizing radiation (IR) are often repaired by this mechanism [4]. It is thought to be the predominant type of repair mechanism of DSBs in mitotically replicating cells and has been studied in greater detail. The hallmark of NHEJ is its ability to ligate non-ligatable DNA ends.
In this mechanism, after the generation of DSBs, a heterodimeric protein called KU comprising of KU70 and KU80 subunits, binds to the ends of DNA breaks and occupies a region of 16-18bp [11]. This heterodimer next recruits its catalytic subunit called DNA-PKcs to the DNA end which displaces the KU dimer to the inside and binds at the extreme ends of broken DNA [12]. This results in the formation and activation of trimeric DNA-PK holoenzyme, a key kinase in NHEJ repair [13]. Once this kinase is activated, it recruits and phosphorylates key DNA damage repair proteins including WRN, which is a 3’ to 5’ exonuclease [14], the Artemis factor, which has specific 5’ to 3’ exonuclease as well as endonucleolytic activity on 5’ and 3’ hairpins and overhangs [15] and the replication protein A (RPA) which binds and subsequently stabilizes single-stranded DNA intermediates formed during DSBs and thus prevents complementary DNA from reannealing [16].
The recruited exonucleases can digest and process the damaged DNA ends for ligation. Next, DNA-PK phosphorylates XRCC4, a binding partner of DNA ligase IV, which is then recruited to the damaged DNA ends. DNA polymerases POLμ or POLΛ are next involved in filling the DNA gaps produced by the damage [17] while XRCC4-Ligase IV complex seals the nicks [18]. DNA-PK also autophosphorylates itself, which causes its detachment from the DNA lesion [19]. A simplified cartoon version of the entire process of DNA repair mediated by NHEJ is depicted in figure 2.
![Figure 2. Double stranded DNA repair via Non-homologous end joining (NHEJ). After DNA undergoes double stranded breaks, it may be repaired in an error prone mechanism via NHEJ. The DNA breaks are first detected by KU subunits which recruit DNA-PK catalytic subunit and assemble DNA-PK holoenzyme. This results in the recruitment of specific endonucleases that carry out DNA ends processing for subsequent ligation, DNA polymerase μ or λ to fill in the gaps and XRCC/LigIV complex to seal the DNA nicks. The Figure is designed on the basis of information from [11, 12, 15]. Figure 2](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-2_550.jpg)
1.2.2. Homologous Recombination repair (HRR)
This mode of DNA repair is referred to as error-free mechanism. DNA damage caused by either IR or replication fork arrest are primarily repaired by this mechanism. The reason why greater repair accuracy is achieved with this mechanism is because of the use of homologous sequences in the genome e.g. sister chromatids, homologous chromosomes or repeated regions, to prime the repair synthesis. For this reason, HRR is most active in late S/G2 phase of the cell cycle [20]. The fact that HRR can use homologous chromosomes as a template for the resynthesis of damaged DNA, wich can result in loss of heterozygosity, is an important feature of HRR repair.
The HRR process is triggered by the generation of DSBs primarily post S phase. This is followed by extensive 5’ to 3’ end processing resulting from the activities of the recruited exonucleases i.e. MRE11, present as a complex in MRE11/Rad50/NBS1, collectively called MRN complex [21] and WRN [22]. Additional roles of MRN complex are also presumed namely: recruitment of the key protein kinase in DSB repair known as ATM, via its NBS1 subunit, which is known to be a substrate for ATM phosphorylation [23] and unwinding DNA via ATPase activity of its Rad50 subunit [24]. After the 5’ to 3’ resection, the exposed single strands recruit RPA protein, which coats the resected single stranded DNA and also recruits RAD51 [25]. RAD51 forms nucleoprotein complexes on RPA-coated single stranded DNA to initiate strand exchange, with the help of its five paralogues, RAD51B, C, D, XRCC2 and XRCC3. The resulting nucleoprotein filament searches the nearby sequences to find a homologous sequence which is later invaded with the help of RAD54, an ATPase related to DNA helicases [26]. This is followed by the recruitment of RAD52 which forms a seven-monomer ring structure at the nucleoprotein filament that can interact with both double and single stranded DNA [27]. Additionally, breast cancer susceptibility genes, BRCA1 and BRCA2 are also recruited to the site, which can act as scaffolding proteins and may also interact with RAD51 and RPA [28].
All these initial events result in stand invasion of the DNA overhangs and pairing with homologous sequences. Using this homologous sequence as a DNA template, DNA polymerase starts filling the gaps of the resected 5’ to 3’ DNA ends of the damaged homologue. At this point, there is a formation of an intermediate DNA structure, called Holliday junction [29]. During HRR, this Holliday junction can be resolved either by disengagement of the two pairs of strands resulting in a non-crossingover mode of HRR, or via its endonucleolytic cleavage mediated by resolvases resulting in a crossover mode [5]. Finally, the DNA ends are ligated. The DNA polymerase and the ligase involved in HRR are as yet unknown. The entire scheme of DNA repair via HRR pathway is illustrated in figure 3.
![Figure 3. Double DNA break repair via Homologous Recombination (HR). After DNA undergoes DSBs, it may be repaired via HR. The DNA with DSBs is shown in black, the DNA from sister chromatid or homologous chromosome is shown in orange while the newly synthesised DNA is depicted by blue dashed lines. In this mechanism of error free repair, the DSBs recruit MRN complex, which has exonuclease and ATPase activity via its MRE11 and RAD50 subunits respectively and which may recruit ATM via its NBS1 subunit. Resection of DNA strand results in recruitment of RPA, which further recruit Rad51, its paralogues and Rad54, together forming nucleoprotein complex at the site of damage. Recruitment of Rad52 commences homology search and strand invasion, which can result in the formation of D-loop structure. Unknown DNA polymerases extend the resected DNA strands using homologous sequences as template which results in the formation of Holliday junction. The Holliday junction may be resolved either in crossingover or non-crossingover fashion and the nicks are sealed by DNA ligases. The figure is designed based on information from [23, 24, 26, 29]. Figure 3](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-3_550.jpg)
1.2.3 Single strand annealing (SSA)
Single strand annealing is a non-conservative process of DNA repair of DSBs. In this process, when DNA breaks are produced, DNA resection takes place most likely by the exonuclease activity of MRN complex. The length of the resected DNA, and hence of the DNA overhangs, depends upon how far up within DNA sequence, homology is found within direct repeats. Once homology with either of the two DNA overhangs is found, strand annealing takes place at those homologous regions, while the rest of the non-homologous sequences are cleaved [5]. Repair via SSA does not require RAD51. However, for the purpose of homology search, heptameric RAD52 ring formation still takes place. ERCC1/XPF nuclease is known to be involved in the endonucleolytic activity [30]. Finally, the gaps are filled with DNA polymerase and followed by ligation of DNA ends. Figure 4 shows the scheme of repair via SSA.
![Figure 4. Double DNA break repair via Single Strand Annealing (SSA). After DNA undergoes DSBs, it may be repaired in a non-conservative mechanism via SSA. MRN mediated resection of damaged DNA ends causes recruitment of RPA, which coat the resulting single strands and recruits Rad52 that commences search for homology sequence in the direct repeats. ERCC1 causes endonucleolytic cleavage following by gap filling and ligation. This mode of repair results in loss of DNA sequence and hence known as non-conservative DNA repair. The Figure is designed based on information from [5, 30]. Figure 4](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-4_550.jpg)
As illustrated in figure 1, cellular responses generated after genotoxic insults not only depend on the type of DNA damage and the associated repair mechanism, but also on the scale or magnitude of a particular damage. Hence, if damage is of a lower scale, DNA damage sensors, e.g. KU subunits, MRN complex or WRN would detect the damage and recruit specialized repair enzymes as mentioned above to efficiently repair the damage. However, if there is an extensive DNA damage, with an impact on the entire cellular physiology, the cellular machinery governing cell-fate decision is mobilized. These signalling responses that are triggered after extensive DNA damage not only activate and recruit repair enzymes, but also give rise to a pathway called DNA Damage Response (DDR) pathway [31]. The most important function of this pathway is to link the signalling generated in response to DNA damage with the cell cycle signalling pathway. Figure 5 represents a simple illustration of this link.
![Figure 5. The DNA damage pathway and its crosstalk with the cell cycle. Lines with arrow heads represent activation while lines with bar heads represent deactivation, dashed lines with arrow heads indicate activation via phosphorylation and dashed lines with bar heads denote phosphorylational inactivation. Grey lines with arrow heads represent dephosphorylation. After DNA damage, P53 and checkpoint kinases Chk1/Chk2 proteins are activated. P53 activation results in concomitant rise in the CIP/KIP proteins, P21 and P27 which inhibit cyclin/CDK complexes, blocking the cell cycle progression which results in G1/S arrest. The activated chk1/chk2 kinases activate Wee1, a protein kinase as well as inhibit the function of Cdc25 group of phosphatases, which normally dephosphorylate and activate cyclin/cdk complexes. The inactivation of cyclinB/CDK1 activity via both phosphorylational inactivation by wee1 as well as inhibition of its activating phosphatases by Chk1/Chk2 result in G2/M arrest. Thus in a normal cell, both G1/S and G2/M checkpoints are functional and may arrest the cell cycle upon DNA damage, to allow time for DNA repair. Source Khalil HS et al. [10]. Figure 5](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-5_550.jpg)
2. Ataxia-telangiectasia mutated (ATM)
The DDR pathway has evolved to be a complex, yet sensitive, highly integrated and interconnected pathway which can trigger cellular responses including DNA repair, cell cycle arrest and apoptosis [32]. Central to DDR is the Ataxia telangiectasia mutated gene that codes for Ataxia telangiectasia mutated protein (ATM), a 370 kDa Serine/Threonine kinase, functioning as a core component of the DDR signalling pathway [33]. ATM belongs to phosphatidylinositol-3 kinase like kinase (PIKK) super family of large proteins having phosphatidylinositol-3/4 kinase (PI3K/PI4K) catalytic domain thus functioning as an important kinase in DDR signal transduction. It acts as a sensor of DSBs and through its kinase function, is responsible for the initiation of a signalling cascade by activating other downstream signal transducers and effector proteins of the DDR pathway. These effectors in turn modulate cell cycle progression, recruit DNA repair enzymes and may also trigger apoptosis if the DNA damage is beyond the repair capacity of the cell. Hence, ATM links responses generated by DNA damage with cell cycle progression and apoptotic pathways. This represents a very important function as in conditions of extensive DNA damage, cell cycle must be temporarily halted to allow adequate time for recruitment of DNA repair proteins and the actual repair of DNA. This ensures the integrity of genomic DNA and overall health and survival of dividing cells and prevents partially repaired DNA to pass on to daughter cells. Alternatively, ATM signalling may lead to permanent arrest of cell cycle, or in extreme cases to trigger apoptosis [34]. ATM and other members of the PIKK family are also involved in DNA replication and recombination, and homologous repair during normal meiotic recombination events [35].
2.1 The ATM gene
The ATM gene is located on chromosome 11 (11q22-23) as determined via linkage analysis [36]. It belongs to the class of housekeeping genes [37]. The full genomic organization along with intron-exon boundaries was determined by [38]. ATM gene consists of 66 exons which span around 150 Kb of the whole genomic DNA, with a coding sequence of 9168bp [39]. The entire ATM contig shows a low GC content of 38.1% [40]. The first two exons are termed 1a and 1b. The initiation codon lies in exon 4 and the last exon which is 3.8 kb is the largest [38]. The first 4 exons of the ATM gene, which lie in the 5-UTR have been found to undergo extensive alternative splicing. Apart from that, the 3.6 Kb long 3-UTR, spreading across the last exon, also possesses alternative polyadenylation sites [38]. This results in generation of different mRNA transcripts having varying sequences and lengths. It has been suggested that the different UTRs within ATM mRNA transcripts may have important regulatory roles via the formation of different secondary structures and varying number of AUG codons [41].
Because of the sheer size of the ATM, special cloning and expression vectors were required for its clonal manipulation. The first successful attempt to clone the whole ATM gene, via positional cloning, dates back to 1995 [33]. Other attempts to clone either its cDNA fragments coding only the kinase domain of ATM [42] or full length ATM [43] quickly followed. Shortly after the successful cloning of the full length ATM cDNA, several groups developed mouse models of a dysfunctional ATM that helped elucidate different functional aspects of ATM [44, 45].
2.2 ATM promoter regulation
ATM shares a bi-directional promoter with another housekeeping gene called NPAT which lies around 0.55kb upstream of the ATM start codon. NPAT is required for progression through G1/S and entry into S phase of the cell cycle and has also been shown to positively regulate ATM [46]. The ATM side of the promoter activity is found to be 3 times stronger than on the NPAT side [47]. This promoter region has been found to be TATA-less, has CCAAT boxes and several other important promoter sites including CREB, SP1, AP-2, GCF [48, 49]. Full sequence of the characterised ATM promoter region and details of its cis-regulatory elements are shown in figure 6.
![Figure 6. ATM promoter sequence and cis-regulatory elements [48]. Figure 6](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-6_550.jpg)
There is some evidence of the existence of another putative promoter region immediately upstream of the first coding exon which also possesses TATA box [50]. This may contribute in the basal level expression of the ATM gene. In the past, several researchers including our group have targeted the ATM promoter region to study its activity or elucidate ATM expression patterns [51-56]. While ATM is generally regarded as a constitutively expressed gene with no major change in its overall expression, it has been found that the promoter activity is tissue specific, and shows an induction of transcription in certain conditions [51, 52].
A higher induction is seen in tissues with lower basal level of ATM and vice versa [57]. Moreover, some tissues show higher protein expression without any changes to mRNA levels suggesting a translational control rather than promoter regulation [58].
2.3 ATM protein: Domains, post-translational modification and expression
ATM is expressed as nuclear 3056 amino acid-long Serine/Threonine protein kinase [43, 59]. The 3-D crystal structure of this protein is as yet, unknown. However through protein sequence analysis and homology determination, several protein domains have been identified in ATM.
The kinase function of ATM is maintained by 350 residues long, PI3K/PI4K domain spreading between amino acids 2712 to 2962. It also contains a FAT domain (name derived from FRAP, ATM and TRRAP) at region 1960 to 2566 amino acids and a C-terminal FAT domain (FATC domain) between residues 3024 and 3056 [60]. Apart from that, a leucine zipper motif between residues 1217—1238 [33, 60] and a ten amino acids long, proline rich c-Abl (a tyrosine kinase) interacting region between residues 1373 – 1382 have been identified [61]. Figure 7 shows the domain architecture of ATM protein along with their associated functions.
![Figure 7. The domain architecture of ATM and the associated functions. The 3056 amino acid long ATM protein has been shown to have a FAT domain, a kinase domain and FATC domain at the extreme C-terminal. The 251 amino acid long, C-terminal kinase domain of ATM classifies it in the PIKK superfamily of protein kinases. Source Khalil HS et al. [10]. Figure 7](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-7_550.jpg)
ATM is a constitutively expressed protein, which is held in an inactive homodimeric or higher order multimeric state. In this form, the kinase domain of one molecule is buried in a region surrounding the residue serine 1981 of the partner monomer. Upon genomic insults e.g. exposure to ionizing radiation, the pre-existing ATM molecules undergo rapid autophosphorylation at residues Ser367, Ser1893 and Ser1981, the last serine being in the FAT domain [62, 63]. This autophosphorylation results in dimer or oligomer dissociation and the release of kinase active monomers [62]. These monomers have exposed N-terminal sequences to bind to their substrates while C-terminal kinase domain for their subsequent phosphorylation. DNA damage-induced full activation of ATM also involves acetylation of Lysine 3016 present in the FATC domain by Tip60 histone acetyltransferase [64, 65]. The complete list of phosphorylated ATM residues determined so far is given in table 2. However, of those listed, only the autophosphorylation sites at Serine 367, 1893 and 1981 have been fully characterised and play a role in ATM activation [62, 63, 66]. The remainder of the phosphorylation sites were identified through large scale proteomic analysis [67-69] and their role in regulating the function of the protein warrants further study.
![Table 2. ATM phosphorylational mapping [10]. Table 2](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Table-2.jpg)
In terms of its expression, ATM is regarded as a housekeeping gene with predominantly nuclear localisation and a steady constitutive expression [70]. ATM maintains basal levels of phosphorylated ATM at serine 1981. ATM expression and localisation in human keratinocytes is shown in figure 8. At an organism level, it is ubiquitous and is expressed in several embryonic and adult tissues [71]. However, the overall expression varies from organ to organ. High expression has been observed in developing nervous system [72, 73], spleen, thymus and testis [53, 71]. Additionally, higher amounts are also seen in those tissues which undergo frequent proliferation and genetic recombinations to ensure the genomic integrity [57, 74].
![Figure 8. Expression and localisation of ATM and phosphor-ATM (serine 1981) in HaCat cells. Cells were grown on poly-L lysine coated coverslips for 18 hours and transfected with N-terminal YFP tagged ATM. 24 hours later, cells were fixed, permeabilized and subjected to immunofluorescence by labelling with anti-phospho-ATM (serine 1981) antibody and Alexa fluor 568 conjugated secondary antibody (Invitrogen). Scale bar represents 8μm. Source - Khalil HS et al. [75]. Figure 8](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-8_550.jpg)
ATM is regarded as the caretaker of the genome [76] and apart from being activated after genomic insults, is also thought to be involved in oxidative stress response [77, 78] (more details in section 5).
In keeping with its role as a constitutively expressed gene, ATM protein level is generally believed to remain constant within the cells. It has been reported that ATM does not undergo any change in its total protein levels after DNA damage, with the only change being dimer to monomer transition and activation of its kinase activity [62, 79]. ATM expression was earlier also reported to remain constant throughout the cell cycle. Furthermore, following genotoxic insults, no major change in its sub-cellular localisation had been observed initially after its discovery [80].
However, few years later, contrary reports emerged demonstrating an alteration in ATM protein expression under certain circumstances accompanied by a corresponding change in ATM activity. In one study, radiation-induced upregulation of ATM in situ and in response to mitogens resulting in increased ATM kinase activity was reported [81]. By contrast, epidermal growth factor was reported to down-regulate ATM at the transcriptional level [55]. Furthermore, we have shown that the total and phosphorylated levels of ATM underwent cell cycle dependent changes in MCF10A cell line (figure 9).

In most of these instances, alterations in the amount of ATM protein resulted in variation in its activity, ultimately impacting cellular sensitivity towards genotoxic agents. Promoter studies of ATM further revealed radiation inducibility [54] and tissue-dependent variation in expression in vivo [53]. In addition recently it has been shown that BRCA1/E2F1/CtIP binding to ATM promoter activates ATM transcription [82]. Finally, recently we have reported a link between ATM activity and its expression via promoter studies [51, 83]. All these later discoveries challenged the earlier belief of ATM gene to be a constitutively expressed gene and indicated the presence of additional mechanisms through which ATM expression and activity could be modulated, apart from the mere dimer to monomer transition event.
2.4. The ATM signalling pathway and its role in DNA damage response
As mentioned earlier, ATM is present at a nodal point in the DDR pathway, and has the ability to trigger variety of cellular responses including DNA repair, cell cycle arrest and apoptosis in nuclear cells. ATM, as a DSBs sensor, not only ensures upregulation of repair enzymes followed by prompt DNA repair, but also signals to a variety of other key proteins with a consequence on cell-fate. This important link between DNA damage, the cell cycle progression and cellular apoptotic machinery is provided by ATM function and is illustrated in figure 10.
![Figure 10. The ATM pathways and their associated consequences on cell-fate. ATM is present at the core of DNA damage pathway, activated upon DSBs and functions via multiple routes. While great deal of cross-talk exists between individual pathways, its major downstream substrates for DNA repair are MRN complex, BRCA1, RAD51and, P53, for cell cycle arrest are SMC1, CIP/KIP family of proteins via P53 and checkpoint kinases, for chromatic remodelling are Chk1 and for apoptosis are c-Abl, P53, Chk2, E2F1, P73 and NFkB. Source - Khalil HS et al. [10]. Figure 10](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-10_550.jpg)
Since ATM function is implicated in a number of responses resulting from DNA damage, it needs to interact in a timely manner with a variety of downstream effectors. These interactions induce various signalling functions such as checkpoint arrest with P53, Mdm2 and Chk2 in the G1 [84-86] damage induced S-phase arrest with NBS1, BRCA1, FancD2 and SMC1 [87-90] G2/M arrest with BRCA1 and hRad17 [91, 92] and apoptosis with E2F1, Chk2, P53, P73 and Bax [91-97]. Table 3 shows a classification of ATM substrates with respect to their roles in influencing different phases of the cell cycle.
![Table 3. ATM substrates in different phases of the cell cycle. Table is based on data from [10, 85- 87, 90]. Table 3](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Table-3_550.jpg)
Once DNA damage causes DSBs, a change in the higher order of chromatin structure is inflicted, because of the unwinding and relaxation of the local DNA super-coil. This topological change in DNA results in the exposure of a variant form of histone H2A, called H2AX [62]. The exposed H2AX is a substrate for ATM and is thought to trigger its activation. Activated ATM and the pre-existing constitutively active ATM phosphorylate H2AX at serine 139, further referred to as γ-H2AX. H2AX is regarded as the earliest substrate of ATM after DSBs and hence γ-H2AX formation represents one of the earliest events after DNA damage [98]. γ-H2AX acts as a docking site for variety of other proteins involved in the DNA damage response pathway which in turn recruit additional ATM molecules to amplify its signal and form discrete ATM foci at the broken DNA ends. Figure 11 shows such DNA damage induced foci formation in a human keratinocyte cell line exposed to a DSB inducing agent Doxorubicin [99]. A number of proteins are involved in the formation of such ATM foci. These proteins include MRN complex (composed of MRE11, RAD50 and NBS1), MDC1, RPA, RAD51, RAD52, RAD54, BRCA1 and BLM1 in the DNA repair component of DDR pathway [100].

The MRE11 component of the MRN complex executes 3’-5’ exonuclease activity, the RAD50 component maintains the broken ends intact and provides ATPase activity, that itself has endonucleolytic activity, while Nbs1 recruits and possibly triggers further activation of ATM molecules [21] which also undergo acetylation by Tip60 [65]. RPA phosphorylation by ATM is thought to divert its role from DNA replication to repair [101]. BRCA1 is an important ATM substrate having multiple functions. It has a role in homologous repair by activating the enzyme Rad51. Its phosphorylation by ATM at residues Ser1423 and Ser1524 triggers its transcriptional activity causing increased transcription of P21 and GADD51, hence having a role in cell cycle arrest as well [102].
BRCA1 is also thought to act as a scaffolding protein for the multiple protein complexes that form discrete foci at the broken ends. Finally, ATM kinase activity for some of its substrate itself is dependent on BRCA1 protein [103].
These initial events lead to generation of pool of active S-1981 autophosphorylated ATM molecules that rapidly accumulate in the nucleus and amplify and transduce the signals to its effector molecules. DNA damage dependent nuclear pATM S1981 accumulation is shown in figure 12. Activated ATM also modulates the cellular machinery that determines the fate of the cell, other than the DNA repair component. ATM links the DNA damage response to cell cycle checkpoint arrest and apoptosis partly via the tumour suppressor protein P53.

ATM phosphorylates P53 at serine 15 that causes its increased stability and activation [84]. ATM also contributes in P53 activation via mdm2 phosphorylation at serine 395 that hinders mdm2 interaction with P53 [86]. Additionally, ATM phosphorylates both checkpoint kinases Chk1 and Chk2 upon DNA damage [104, 105], which in turn phosphorylate P53 on serine 20 and contribute further in its stabilization [106]. Once P53 is activated, it acts as transcriptional factor for important proteins including inhibitors of CDKs e.g. the CIP and KIP family of CDK inhibitors, P21/waf1 and P27. As shown in figure 5, these bind and inhibit CDKs, an event which causes cell cycle arrest at G1/S phase. P53 also activates GADD45 via which, it links itself to ATM induced DNA repair. Another downstream target of ATM implicated in S-phase arrest is Fanconi anemia group D2 protein (FANCD2) phosphorylated at Ser222 [88]. P53 is also responsible for causing apoptosis in ATM dependent manner [94].
Apart from the above-mentioned role of the checkpoint kinases Chk1 and Chk2 in P53 stabilization, these effectors also phosphorylate Cdc25 phosphatases. Chk2 phosphorylates Cdc25A phosphatase at serine 123 which causes its inactivation rendering it unavailable to dephosphorylate and activate CDKs, which otherwise cause cell cycle progression. This results in checkpoint arrest mainly at the G1/S phase [104]. Chk1 regulates the activities of Cdc25B by its phosphorylational inactivation and causes cell cycle arrest mainly in G2/M phase [107]. Furthermore, Chk1 also phosphorylates Cdc25C on serine 216 that serves as a binding site for 14-3-3s protein, which sequesters it from nucleus [108] rendering it unavailable to dephosphorylate and activate cdk-cyclin complexes important for mitotic entry (fig. 5).
As illustrated in figure 10, ATM also plays a pivotal role in apoptotic induction both in P53 dependent and independent manner. Firstly, ATM can phosphorylate E2F1 on serine 31, which results in its increased stability and can cause apoptosis in P53 dependent manner [97, 109] as well as in an independent manner [110]. E2F1 can also undergo Chk2 dependent phosphorylation after DNA damage and trigger apoptosis [111]. Additionally, ATM dependent apoptosis can also be induced via Chk1 [112]
In a P53 independent mechanism, ATM can phosphorylate and activate c-Abl tyrosine kinase which via P73 dependent mechanism can induce apoptosis following extensive DNA damage [113]. Finally, ATM can also modulate apoptotic pathway following DNA damage, by stabilizing nuclear factor kappaB (NFkB) (reviewed in [114]).
2.5. Protein trafficking during DDR
There is a vast amount of literature that aims to describe the functioning of DDR pathway from sensing the DNA damage to the recruitment of effector molecules and triggering the cellular machinery involved in cell-fate decision making. While our understanding of the causes of cellular stresses that lead to DNA damage and their consequences for key events like transcriptional changes, post translational modification and the type of protein repair complexes that are assembled, has improved over the years, there are still gaps in our knowledge of DNA damage induced protein trafficking responses. However, only in the last decade, it has been realised that eukaryotic protein trafficking machinery, apart from facilitating the processing, modification and secretion of nascent proteins, is also actively involved in different regulatory modalities and co-ordination of cell signalling and forms active component of key cellular decision making processes e.g. apoptosis, proliferation and cell cycle events.
Protein trafficking represents a very important constituent of eukaryotic cellular physiology. It is not only used to distribute newly synthesised and correctly folded proteins to their final destination, but also to facilitate transport of mature proteins from one sub-cellular compartment to the other for their proper functioning. In terms of DDR signalling, these trafficking events may take place to either provide association of DDR enzymes with their substrates, to sequester/bind a DDR transcription factor from/to its target promoter, or to prevent/allow complex formation between two proteins which would trigger or alter DDR signalling. Furthermore, within signalling pathways, intracellular protein trafficking events can also not only influence the overall signalling efficiency of a particular pathway, but can cause distinct compartmentalization of a multifunctional protein, a phenomenon that can lead to a bias in its downstream signalling event.
In terms of DNA damage, the most critical function of the protein transport system is to concentrate the repair protein complexes at the site of DNA damage. This is expected to result in the translocation of multitude of proteins to the site of DNA lesion. This is a rapid process as indicated by the fast kinetics of γ-H2AX phosphorylation and the resulting DNA foci formation, estimated to occur within minutes of DNA damage [115]. For this reason, protein trafficking events within the nucleus in response to DNA damage has received greater attention.
The role of protein trafficking in cell cycle progression is provided by a typical example of phosphorylation of Cdc25C by checkpoint kinases. Cdc25C phosphorylation at serine 216 by Chk1 creates a binding site for 14-3-3, which mediates its transport to the cytoplasm, sequestering it from its substrate, CyclinB-Cdk1 to prevent cell cycle progression [108]. Another example is the controlled nucleo-cytoplasmic shuttling of E2F1 protein that is required for the progression of cell cycle [116]. Illustration of the importance of protein trafficking in the DNA repair component of DDR is provided by BARD1 protein, which is retained in the nucleus by BRCA1 resulting in DNA repair. In the absence of BRCA1, BARD1 is transported to cytoplasm where it triggers apoptotic signalling [117].
P53 is widely characterised to shuttle between nucleus and cytoplasm which is influenced by cell cycle progression and DNA damage (reviewed in [118, 119]). While in the cytoplasm, P53 may form complex with antiapoptotic protein Bcl-xl preventing apoptosis, any nuclear P53 induction will trigger apoptotic proteins e.g. PUMA which itself can traffic to cytoplasm and release P53 from the Bcl-xl complex. This leads to mitochondrial membrane permeabilization resulting in apoptosis [120].
Secondly, while nuclear translocation of P53 triggers its proapoptotic transcriptional function upon treatment with anticancer DNA-damaging agents, the same molecule is transported to mitochondria upon treatment with a tumour promoting agent, where it inhibits the activities of tumour suppressor, Manganese superoxide dismutase [121]. P53 is also known to translocate to mitochondria following radiation or camptothecin treatment, while APE-Ref1 and BAX, after treatment with H2O2 and etoposide respectively [122-124]. Other proteins involved in DDR e.g. BRCA1 [125], Chk1 [126], Cdc25A [127] and BID [128] show DNA damage induced nuclear export, while still others e.g. Mdmx [129] and Optineurin [130] undergo nuclear import following DNA damage. A vast majority of proteins including the DDR kinases show intranuclear relocalisation upon DNA damage e.g. DNA-PK [131], ATR [132], Mdm2, [133] and p14ARF [134].
The ATM kinase was first regarded as a nuclear protein that was believed to function mainly in the nucleus as a critical DNA damage sensor [71]. Discoveries of ATM’s other functions beyond its well characterised role as a nuclear DNA damage sensor kinase was expected to require some degree of sub-cellular ATM trafficking. In terms of ATM’s role in cell proliferation, it was shown that upon double-stranded DNA damage, a fraction of nuclear phosphorylated ATM underwent protein trafficking to the cytoplasm where it activated NFkB [135]. On the other hand, it was also shown recently that damage induced DDR was accompanied by nuclear import of cytoplasmic pATM following UV treatment [136].
Interest in characterising the extra nuclear role of ATM was triggered by the increasing understanding of the disease phenotype of Ataxia Telangiectasia (A-T) patients, caused by deleterious mutations in ATM gene (Refer to section 3). ATM deficiency in patients leads to cancer susceptibility, weak immune system, hypersensitivity to ionizing radiation and sterility while A-T cell lines undergo radio-resistant and damage prone DNA synthesis and abnormality in cell cycle checkpoint arrest (more details of the A-T disease are given in the following section). While these aberrations can straightforwardly be attributed to the role of ATM in DDR, other neurological disease phenotypes e.g. ataxia, speech defects and abnormal body movements or cellular defects e.g. cytoskeletal abnormalities, plasma membrane defects and high levels of trophic factors requirement for growth cannot all be explained through ATM’s role as a nuclear DNA damage sensor.
These initial questions led to key discoveries, which supported the role of ATM kinase further than its conventional role in the DDR pathway [45, 87,137, 138]. Table 4 lists the localisation and associated functions of ATM in different sub-cellular compartments reported so far, other than its nuclear function.
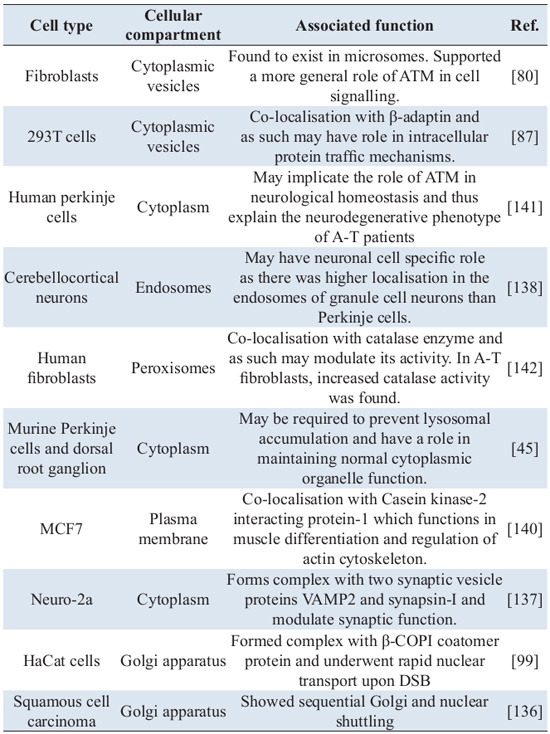
It is imperative that such differential localisation of the same molecule in different cellular compartments involves rigorous protein trafficking events. The fact that ATM has been shown to interact with karyopherins [139], found to co-localise with β-adaptin [87], is recruited from nucleus to the plasma membrane [140] and the recent discovery of sequential localisation of pATM in Golgi apparatus and nucleus in normal human primary keratinocytes as well as in squamous cell carcinoma cell lines after either UV treatment [136] or chemotherapeutic drug [99] also supports the idea that ATM undergoes extensive protein trafficking.
3. Ataxia telangiectasia (A-T) disorder and clinical symptoms
The importance of the role of ATM is highlighted by its functional loss in the disease, Ataxia Telangiectasia (A-T) also known as Louis-Barr syndrome. A-T is a rare autosomal recessive multisystem disorder caused by hereditary mutations in the ATM gene. It is the most common recessively inherited type of cerebellar ataxia in small children with a frequency of 1 in 50,000 live births [143].
The disease is characterised by ataxia, referring to uncoordinated body movements and telangiectasia, which means enlarged blood capillaries that can be seen under the skin (fig.13). Other characteristic features of A-T include immune deficiency, hypersensitivity to ionizing radiation, and a predisposition to certain cancers [144]. Despite the heterogeneity in the clinical presentation of the A-T syndrome resulting in the initial finding of at least 4 complementation groups [144], the linkage analysis showed that they all map to the same genomic location on chromosome 11 (11q22.3) containing the ATM gene [37].
![Figure 13. The Ataxia Telangi-ectasia (AT) disease phenotype. Early symptoms include progressive cerebral ataxia causing defects in motor skills manifested in early childhood. This is followed by the formation of telangiectasia, immune defects and predisposition to malignancy later in life and sterility in adulthood. Patients show severe radiosensitivity whereby normal doses of chemotherapy, to cure cancer caused by AT disease, to be lethal. Source: [155]. Figure 13](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-13.jpg)
Heterozygous carriership of ATM seems to be very common in humans, the estimates varying between 1.4 % and 2.2 % of the general population and even more common (up to 12.5 %) in populations with a marked founder effect [145-147]. A-T patients from nonconsanguineous families are usually compound heterozygotes. Carriers of defective ATM alleles, although generally considered asymptomatic, have been found to have an increased risk of death at any age due to all causes including cancer and ischemic heart disease [147].
3.1 Neurological features
Most common neurological disorder is progressive cerebellar ataxia, which may initially be misdiagnosed as cerebral palsy of the ataxic type. It is apparent as early as the first year of a child. Truncal and gait ataxia are slowly and steadily progressive. This leads to wheelchair confinement in the teenage years. Progression of neurological degeneration continues in the adult life without actual mental retardation. Thus, typical A-T patients are of normal intelligence, although abnormalities in the motor skills make normal learning programs difficult. Abnormalities in the neuronal cells of A-T patients include degeneration of Purkinje cells and thinning of the granule cell layers and some abnormalities in olivary nuclei and medullary tracts [144].
3.2 Telangiectasia
Telangiectasia is the dilation of blood vessels that are visible through the skin. Usually telangiectasia appears 2-4 years of the manifestation of neurological abnormalities, similar to the ataxia, appearance of telangiectasia is progressive as well. First signs of telangiectasia appear mostly in conjunctiva, followed by appearance on ears, over nose and behind the knees. These dilated capillaries are normally not associated with bleeding. The absence of telangiectasia does not exlcude the diagnosis of A-T.
3.3 Radiosensitivity
Radiosensitivity of A-T patients towards ionizing radiation is understandable owing to the prominent role of ATM in the desensitization towards genotoxic insults. Hence, A-T patients with cancer are extra sensitive to those doses of ionizing radiation, which are otherwise employed for non-A-T cancer patients and this sensitivity is to a life threatening level.
An interesting observation in some A-T patients is the loss of the distal 11q region of the chromosome that harbours key DNA damage response genes including ATM, MRE11, Chk1 and H2AX in several haematological malignancies and solid tumours. At a cellular level, A-T cells, which lack detectable ATM, are unable to detect levels of DNA damage which usually result in activation of the DNA repair machinery, causing sustained DNA damage. Also, they cannot induce cell cycle arrest, resulting in replication of damaged DNA and multiplication of errors until the burden of damage becomes too severe for the genome. In both cases, the cell is directed to a suicide route. Apart from radiosensitivity, A-T cells have abnormal telomere morphology and genomic instability [148].
3.4 Cancer predisposition
Cancer predisposition is one of the major complications in A-T with the homozygous A-T patients having a life time risk of 30-40 % [149]. The correlation between ATM and some forms of cancers have been long established with cancer being the most frequent cause of death of A-T patients. This link was first observed by Reed WB and colleagues in 1966 who looked at incidences of familial and sporadic cancers in homozygous A-T patients as well as in heterozygous carriers [150]. Numerous statistical studies have been undertaken to form a broader picture to generate a correlation between A-T causing mutations and incidences of different kinds of cancers [39, 151]. There are several reports that attempted to demonstrate links between heterozygous and homozygous A-T patients and incidences of familial and sporadic cancers. Such correlations have been found in variety of tumours to different degrees. This degree of correlation is found to be dependent on the type of ATM mutations e.g. missense or nonsense variants and also the nature of mutations (germ line or somatic A-T).
In line with the finding that A-T patients have a weak immune system, the most common malignancy linked with A-T patients is that of immune system (Leukemias and Lymphomas) with the most common being non-Hodgkin’s lymphoma (~45%) [152] followed by acute lymphocytic leukemias (~20%) [153]. This link has further been demonstrated by generating knock out mouse models of ATM shown to be highly susceptible to sporadic lymphoid malignancies owing to impairment of the V(D)J recombination in such ATM deficient mouse lymphocytes.The lymphomas in A-T patients tend to be of B-cell origin, whereas the leukemias are usually of T-cell origin. While cancer of the immune system is more common in the early lives of A-T patients (mostly occurring in first 15 years), older A-T patients are at a greater risk of developing a variety of solid tumours including gastric, breast, medulloblastoma and basal cell carcinoma [74]. Furthermore, the role of heterozygous carriership of defective ATM alleles has been clearly demonstrated in a proportion of familial breast cancer and colorectal cancer cases as well [39, 154].
4. Role of computational modelling in deciphering DDR pathway
Over the past decade, tremendous progress has been in producing qualitative signalling data that has improved our qualitative understanding of the signal transduction pathways, their molecular mechanisms, activity, regulation and the associated outcome on cell-fate. While this has helped us immensely in conceptualising intervention strategies in diseased states, the decision making property of these pathways may involve oscillations and concentration thresholds that require a quantitative approach, calculations and numeral analysis for data collection and interpretation.
A prerequisite of a molecularly targeted anticancer approach is a detailed understanding of the underlying mechanisms contributing in tumour development. This involves changes in both the types and extents of molecular interactions governing key process in a cell that may predispose it to cancer. Additionally, a thorough understanding of the cell microenvironment, proliferation, growth and stress signals are vital before a successful anticancer approach could be devised. Failure to do so is one reason why theoretically, very effective anticancer strategies could be conceptualised, while they still fail to actualise. Owing to an intricate nature of context dependent signalling networks, high degree of cross-talk and pathway choices and insufficient quantitative data of signalling dynamics, a clear inter-relationship among cellular signalling to cellular response to its ultimate consequence on cellular phenotype has not been fully determined. Until recently, strategies for therapeutic intervention via the modulation of key molecular pathways targeted individual components of the global regulatory network. This was based on the assumption that different pathways transmit signals through independent mechanisms. However, with the advent of new molecular techniques and bioinformatic tools, remarkable progress has been made and new insights have been gained into the nature of the intracellular signalling networks.
It is now known that numerous key proteins have multiple functions in different pathways and that the downstream signalling choice of a particular protein depends on a number of variables. ATM is central to a number of key pathways as mentioned before. These pathways act in concert in order to phosphorylate P53 at a number of sites, some having overlapping functions and others being specific for a particular response. In this micro-environment, the effect of manipulation of DDR in general or inhibiting ATM protein specifically is not fully predictable especially owing to the fact that ATM function can contribute both in numerous physiological process as described in other sections of this book (see section 5). Prediction is further complicated by the finding that ATM may self-regulate its own protein levels [10, 51, 83].
In this complex and unpredictable scenario, the efficacy of potential ATM inhibitors would have to be assessed in terms of the effects it exerts, not only on ATM activity and its immediate substrates e.g. ATM→pATM→pP53→DNA repair, but on a number of connected key proteins in the DNA repair, cell cycle, oxidative stress, protein degradation, metabolic signalling, proliferative and apoptotic pathways and their associated consequence on cell-fate. It is increasingly becoming obvious this complexity in determining the absolute effects of drug intervention necessitates parallel theoretical and experimental considerations. This could be best addressed by employing a systems biology approach to the problem, involving mathematical modelling of biological processes [156-160]. To fulfil this, there is a need for collecting quantitative dynamic information of DDR signalling proteins at high temporal resolution and identify how its sub branches e.g. those controlling DNA repair informs cell cycle regulatory network, or how the energy producing pathways are influenced by apoptotic signaling and so forth [156, 161].
Mathematical model construction based on information such as above would not only provide novel insights into how thresholds, localisation and specific interactions of proteins within DDR signalling are triggered and regulated during the course of genotoxicity, but would also help establish maps of network interactions that would further aid in identifying spatio-temporally regulated critical links and their contributions in pathways responsible for generating a specific cellular response during a specific treatment regime. Once such critical signalling links are established in cancer cells, these could be exploited to devise treatment portfolios to achieve targeted cellular sensitivity. Such a model could be capable of predicting the global phosphorylation status of important proteins e.g. ATM, ATR, P53, E2F1 and BRCA, the activity levels of checkpoint kinases e.g. Chk1 and Chk2, a balance between the action of DDR associated kinases and phosphatases , pro and anti-apoptotic proteins, and their outcome on cellular fate. Also, such a model could be able to provide for varying levels of drug input over variable time courses as well as being capable of incorporating combinations of intervention strategies e.g. involving both ionizing radiation and chemical carcinogens in conjunction with ATM or other DDR protein inhibitors.
In the past, several researchers have targeted a variety of dynamical processes within biological systems for their computational analysis ranging from studies into parallel reaction pathways [162], HBV infections [163] and in-silico analysis of Sar-CoV [164]. In terms of DDR signalling pathway, most of systems biology studies have been undertaken to examine and model the oscillatory patterns of DDR protein induction following DNA damage in light of the known biological insights [165-168] with some reports focussing on elucidating the role of such oscillations in determining cell fate i.e. DNA repair, cell cycle arrest or apoptosis [169-172].
Attempts have also been made to decipher G1/S [173-174] as well as G2/M checkpoint engagement upon genotoxic insults [175-176] and parallel cell cycle analysis of normal and cancer cells using a Systems Biology approach [157-160]. Recently, several system biology attempts to elucidate the mechanism of drug resistance to chemotherapy in different cancer patients have been informative and uncovered underlying pathway alterations driving such drug resistance [177-178].
Systems biology approach for cancer is a new idea and is still in infancy. The above research attempts for testing experimental hypothesis have made it clear that computational modelling and systems biology strategy works best if it is integrated and made an integral component of the experimental investigation. Systems biology is a two-way information exchange between biology and mathematical modelling. The scheme of this relationship is depicted in figure 14 where phase I starts with a biological investigation of a problem, data generation and data mining. This data is fed into a molecular modelling system, where it not only enables visualization of the biological networks in a more understandable and interpretive manner, but also allows for its validation, biological appropriateness and suitability and potential for realising biologically observed phenomena. In phase II, data simulation takes place and the model identifies key variables, generates novel hypothesis, recommends technology development for experiments, and proposes areas of interest for further experimentation. The hypotheses generated are further tested experimentally taking into account the recommendations of variables made in phase II. The data or novel insights obtained are further fed back to the model to revise or improve the predictive power of the model and generate further hypotheses and further recommends technology development for future experimentation in phase-III .

In conclusion, previous mathematical modelling attempts have shown that the design and construction of a deterministic mathematical model of the molecular interactions that underpin the DDR require new kind of time series quantitative data. This data must be consistent, have high temporal resolution and spatial consideration and provide the kinetic parameters of larger number of different proteins involved in a single pathway. The mere descriptive nature of the information regarding key cellular pathways has so far limited their use in mathematical and computational modelling applications, such as those identified in Phase I of the modelling scheme (fig. 14), which could potentially identify new therapeutic targets and help us devise treatment regimens. While qualitative data are easily available, quantitative data pertaining to key signalling molecules that would allow speedy calibrations and provide kinetic parameters for the construction of mathematical model is still scarce.
5. Beyond DNA damage repair: ATM’s role in oxidative stress and cellular metabolism
While ATM activation and function has primarily been characterised in response to genotoxic challenge, growing body of evidence suggests its functional importance in pathways quite distinct from DNA damage and repair. This enables ATM to participate in diverse set of physiological processes involving metabolic regulation, oxidative stress, transcriptional modulation, protein degradation, cell proliferation and cancer.
It has been demonstrated for quite some time that in addition to DNA damage, ATM may also be activated under oxidative stress and aging in the absence of DNA damage. [179]. This has been shown both in in vitro as well as in in vivo studies [180]. The idea that ATM may function in oxidative stress response following oxidative damage of macromolecules other than the DNA, came from the frequent observation of defects in cellular antioxidant systems, elevated reactive oxygen species (ROS) and deregulated levels of ROS scavenging enzymes at cellular level, animal models and AT patients. It has been demonstrated that cells lacking ATM had an impaired capacity to synthesise glutathione [181]. This finding had clear implications of ATM in redox signalling pathway. This led to further in vitro investigations in the coming years that attempted to discern ATM functioning in oxidative stress response which supported ATM’s role in these domains [182, 183]. In another report, study of the anti-oxidant capacity of ten A-T patients as compared to age matched controls showed a reduced antioxidant capacity and altered levels of reactive oxygen species (ROS) scavenging enzymes [184].
In the same year, in an in vivo study, it was demonstrated that mice lacking the ATM gene showed extensive oxidative tissue damage in organs most affected by the A-T disease thus providing a mechanistic basis for A-T disease phenotype [185]. Interestingly, the oxidative stress induced ATM activation without DNA damage. For example, ATM activation and its cytoprotective role in oxidative damage was reported following treatments with H2O2 and C2– Ceramide, agents that induce oxidative damage of cellular macromolecules, without causing chromosomal breaks [186]. A year later, two further in vivo studies reported an altered redox state in brains of mice that lacked ATM gene [187, 188]. It has been hypothesised that the high oxidative stress, as suggested by altered levels of key enzymes, catalase and superoxide dismutase, may explain the neuronal degradation observed in A-T patients [187]. Further reports of in vitro elucidation of ATM signalling in oxidative stress revealed that cells lacking ATM also had defects in oxidative stress induced G1 and G2 checkpoint function [189].
In keeping with ATM’s role in oxidative stress, further studies showed that the phenotypic abnormalities seen in ATM null cell lines could be rescued by the addition of antioxidants, thus demonstrating that at least a part of the cellular defects seen in ATM null genotypes are because of accumulation of oxidative damage [190-192]. Thus, as mentioned, sufficient evidence was provided to support the role of ATM in other important physiological processes in addition to being a central component in DDR.
However, a key question that remained unanswered well until couple of years back was how ATM functions as a sensor oxidative stress and gets activated not involving DNA damage? The mechanism was thought to be more complicated and unique as it was reported that oxidative stress caused disruption of DNA binding activity of MRN and hence ruled out MRN mediated activation of ATM [77]. The answer was provided by two reports in 2010 [77, 78]. According to these reports, intra-cellular oxidative stress induced ATM activation by modifying its cysteine residue at position 2991. This caused formation of disulphide bond between two ATM monomers resulting in formation of active ATM dimers. This was a very novel and striking discovery that, contrary to the DNA damage induced ATM dimer to monomer activation, occurred in the reverse order. Mutation of this cysteine to alanine residue disrupted oxidative stress-induced ATM activation but retained its DSB dependent activation. Interestingly, in vitro experiments showed that oxidative stress-induced ATM activation did not require DNA, MRN and the presence of serine 1981. While treatment with a pro-oxidant H2O2 resulted in ATM mediated Chk2 and P53 phosphorylation, it did not cause γ-H2AX formation (marker of DNA damage), or phosphorylation of KAP1 protein, a phosphorylation event that occurs following DNA damage [77, 78]. This was a key observation showing that oxidative stress or DNA damage induced ATM activation may have a complicated interplay and lead to a different subset of substrate activation. If such substrates, which are uniquely activated either by oxidative stress or DNA damage are identified, this could translate into development of biomarkers that can report and distinguish oxidative stress induced apoptosis from DNA damage induced apoptosis or other events.
Nevertheless, making a distinction between these two events is complicated by the finding that oxidative stress itself can cause DNA damage and there is a great deal of cross talk and overlapping function of proteins. For example, 700 proteins have been identified that responded to ATM activation following DSBs, not all of which were known for functioning in DDR pathway [67]. Figure 15 illustrates most of the oxidative stress induced ATM pathways leading to removal of ROS or alteration in metabolic pathways that normally produce ROS.
![Figure 15. Oxidative stress induced ATM signalling pathway. ATM is activated following generation of ROS via an active dimer formation involving cysteine 2991 of ATM protein. Activated ATM may upregulate p21 levels via P53, which results in nuclear accumulation of NRF2 that induces antioxidant response. ATM can also result in NRF2 activation via PKCδ. Following oxidative stress or insulin treatment, ATM also activates AKT by inducing its phosphorylation at serine 473. Activated AKT triggers transport of cell surface glucose transporter 4 (GLUT 4) which results in uptake of glucose, activation of Glucose-6 phosphate dehydrogenase that generates NADPH, an important cofactor in the antioxidant pathway. ATM also directly phosphorylates Hypoxia inducing factor α, which via indirect route causes inhibition of mTOR. Furthermore, direct and indirect activation of AMPK by activated ATM results in activation of TSC2, which also inhibits mTOR. Inhibition of mTOR not only halts cellular proliferation under oxidative stress, but also results in inhibition of pathways that carryout oxidative metabolism leading to lowering of the oxidative burden on the cell. The figure is designed on the basis of data from [193-197]. Figure 15](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-15_550.jpg)
The above text described in detail the mechanism and pathways of ATM signalling in oxidative stress. Apart from that, it has been shown that ATM may have a more general role in other key physiological events as well, e.g. cellular homeostasis, mitochondrial function [198], metabolic control [199] protein degradation and sumoylation [200] and hypoxia [201]. The fact that AT patients have an increased risk of developing type 2 diabetes because of Insulin resistance and glucose intolerance suggested ATM’s role in Insulin signalling and glucose uptake [202]. The discovery of ATM’s role in regulating Insulin like growth factor 1 receptor [203] and ATM dependent AKT activation following IR or insulin treatment [195] that results in glucose uptake not only served to explain disease phenotype of A-T patients but also supported ATM’s role in metabolic signalling pathways. Other studies supporting ATM function in cellular metabolism reported the involvement of ATM in pentose phosphate pathway that led to generation of Glucose-6-phosphate mediated NADPH [196]. One study reported that a mutant P53 devoid of ATM phosphorylation site not only resulted in elevated ROS, but also led to insulin resistance and glucose intolerance, defects that could be alleviated by the addition of antioxidants [204]. ATM mediated AKT activation mentioned above also results in activation of mTOR signalling that carries out oxidative metabolism resulting in ROS generation. In terms of its modulation of translation, ATM has been known to phosphorylate 4EBP1 protein on serine 111 following Insulin or IR treatment.
This phosphorylation inactivates this protein which otherwise represses the translational initiation factor, ELF4E, which then can initial translation (Yang DQ et al., 2000). Moreover, in addition to its role in cellular proliferation and survival via the AKT pathway, ATM signals in cytoprotection and inflammation pathway through NFkB activation. This represents cytoplasmic function of ATM [205]. Activation of AMPK by ATM via LKB1 dependent [193] and independent mechanisms [206] further establishes ATM’s role in cellular homeostasis and metabolic signalling. Under energy crisis, cells may accumulate ROS because of depletion of NADPH.
Accumulated ROS may activate ATM that further activates AMPK which shuts down energy consuming pathways (proliferation, DNA synthesis etc) and triggers catabolic and energy generating pathways (e.g. autophagy and mitochondrial biogenesis) as well as NRF2 activation to combat ROS. Hence, while Insulin treatment causes ATM mediated AKT activation and pro-survival signal, upon IR or IGF1 treatment, ATM may activate AMPK signal that causes TSC2 mediated mTOR inhibition and cellular arrest. This establishes ATM’s role in a critical decision making machinery that is driven by energy requirements of the cell and total ROS levels. A simplified version of the signalling pathway that balances ATM activity between pro-survival and energy consuming processes that may result in ROS generation and cytostatic and energy yielding pathway that scavenges for ROS is presented in figure 16.
![Figure 16. Role of ATM in balancing energy consuming and energy yielding pathways. Following activation upon Insulin treatment or IR, ATM may activate AKT pathway which further activates mTOR. Activated mTOR inhibits autophagy and carries out oxidative metabolism by utilising cellular energy, which may generate ROS. On the other hand, same treatments can result in ATM dependent activation of AMPK pathway, which yields energy by generating ATP, triggers autophagy and scavenges for ROS. The energy state and degree of ROS may determine the type of pathway activated downstream of ATM. Figure is designed on the basis of data from [193, 195]. Figure 16](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-16.jpg)
Another manner by which ATM regulates the metabolism, mitochondrial function and angiogenesis is via its modulation of hypoxia inducing factor 1 (HIF1). As shown in figure 15, under hypoxic conditions, ATM phosphorylates HIF1, which is implicated in the above mentioned cellular processes. HIF1 is also known to modulate activities of a protein called REDD1. This protein causes sequestration of 14-3-3 proteins from TSC2 proteins, thus releasing TSC2 from inhibition and allowing it to inhibit mTOR. Hence ATM can modulate metabolic pathways either by modulating mTOR activities via AKT and AMPK or through HIF1 activation [197].
6. Novel perspectives: Targeting ATM pathway for therapeutic intervention in cancer
Decades of research efforts in attempts to elucidate DNA damage response pathways have added significant knowledge to our understanding of the mechanisms involved in carcinogenesis, DNA damage and repair pathways, and induction of apoptosis. Scientists have described cancer as the disease of the cell cycle caused by frequent mutations in cell cycle regulatory machinery, proliferative signals and DNA repair enzymes. This definition classifies all the proteins functioning in DNA repair, cell cycle progression, division, arrest and apoptosis as critical determinants of not only the cancer phenotype, but also of the therapeutic approach that is devised to combat it.
Researchers have realised that cellular sensitivity to genotoxic agents in course of cancer therapy could be achieved by modulating the function of key proteins involved in DNA repair, cell cycle arrest and apoptotic mechanisms. We now know that the efficacy of the gold standard treatments of cancer involving chemotherapy and radiotherapy can be further controlled and fine-tuned if it is coupled with modulation if the activities of the proteins involved in the above mentioned processes. The idea is based on the fact that during genotoxic treatments of chemotherapy and radiotherapy, cells respond by rapidly upregulating expression and activity of DNA repair and cell cycle regulating proteins. The increased activity of these proteins will render such cells resistant to genotoxicity owing to rapid repair of the cytotoxic DNA lesions and thus hinder the cellular sensitivity process and ensure survival. In such circumstances, greater dose of genotoxicity would be required for effective killing of these cells. However, increasing the dosage will have increased cytotoxic effects on the surrounding normal cells and hence cause more side effects of therapy.
On the other hand, if the activities of proteins involved in cell cycle arrest and DNA repair are inhibited prior to genotoxic treatment, greater cellular sensitivity towards genotoxic agents during treatment might be achieved. Hence, one important therapeutic strategy for cancer is the identification of important DDR protein inhibitors, which could effectively inhibit the type of DNA repair machinery that is activated and required by cell after a particular genotoxic treatment regimen. Discovery and characterization of such inhibitors would not only help to elucidate the function of the relevant proteins but could also identify therapeutically relevant targets.
After validation of specificity and activity of such inhibitors, the next step is to characterise its overall consequence for cellular fate in a context dependent manner. This characterisation is important because specific inhibitors might still have a wide spectrum of effects on cells. This is because proteins participating in DDR and other interconnected pathways for example cell cycle, DNA damage, oxidative stress and metabolism, have high degree of cross talk and overlapping functions. It is further complicated by the fact that many signalling molecules alter substrate preferences and hence downstream signalling by variables such as energy state of the cell, presence of growth factors, phase of the cell cycle, extent of DNA damage and so forth. Hence context dependent treatment regimens have to be formulated and proven to be effective in cellular sensitisation in a well characterised manner. Having established that, the next key consideration is the selectivity of the cytotoxic action towards cancer cells sparing the surrounding normal cells. Such selectivity could be achieved by identifying key differences between normal and cancer cells and then basing the therapeutic approach on exploiting those differences.
Cancer cells usually acquire superior epigenetic and genotypic changes that may either enhance efficiency of DNA repair machinery, handicap the apoptotic pathway, or disrupt cell cycle checkpoint mechanism all translating into better survival phenotypes and signalling advantages thus rendering cancer cells more accomplished and independent in ensuring their survival in emergency situations e.g. during chemotherapy, as compared to their normal counterparts. Additionally, such cells usually evolve overlapping molecular pathways that can overcome physiological barriers and generate the same signalling response as those that are targeted by a drug in a particular treatment regimen. Due to this, a prerequisite of employing an effective molecularly targeted anticancer therapeutic approach is a full elucidation of its complex molecular signalling network driven by these genetic and epigenetic alterations. Such critical molecular detailing that may be unique for different kinds of tumours may identify genetic weaknesses that would not only serve as a biomarker for distinguishing cancerous cells from a heterogeneous population of cells, but also for it to be targeted for sensitization. This is illustrated in the following example:
Cancer cells often have mutations in the cell cycle regulating proteins and DNA repair abnormalities. Such aberrations in vital cellular functions can potentially make cancer tissues more susceptible to certain therapeutic interventions as compared to their normal counterparts. For example, in more than 50% of all cancers, P53 mutations have been found [207]. This renders the cancer cell incapable of executing the P53-mediated G1/S cell cycle arrest and the repair of DNA upon genotoxic insults. The only checkpoint that is active in such cancer cells is the G2/M checkpoint, where the damaged cells are still arrested and repaired, enabling them to survive. Therefore, abrogation of this G2/M checkpoint in cancer cells for example via ATM inhibition (ATM → Chk1/Chk2 → Cdc25A/B/C → G2/M arrest link) would disrupt the only functionally available G2/M checkpoint in cancer cells and devoid them of any checkpoint arrest and hence sensitise the cells against genotoxic agents (reviewed in [208]). On the other hand, the surrounding normal cells would be less affected by this treatment as they retain a wild type P53 that can still undergo G1/S arrest. This scenario is illustrated in figure 17. This approach provides an opportunity for targeted cellular sensitivity based on functional inhibition of ATM kinase.
![Figure 17. The principle of targeting ATM pathway for achieving cellular sensitivity for therapeutic intervention in cancer. Lines with arrow heads indicate activation, while lines with bar heads indicate inhibition. Red arrow indicates DNA damage caused by radio or chemotherapy while red line with bar head indicates therapeutic inhibition of ATM. Cancer cells with P53 loss of function mutation have a dysfunctional G1/S checkpoint whereas the G2/M checkpoint may still be functional. When cells are exposed to genotoxic agents, DDR pathway is activated which will cause G1/S arrest via P53 pathway and G2/M arrest via checkpoint kinases Chk1 and Chk2 pathway. Normal cells can be arrest in either of these pathways allowing themselves time for DNA repair. However, cancer cells can only be arrested in G2/M pathway via ATM→Chk1/Chk2 pathway. Disruption of G2/M checkpoint by way of ATM inhibition would result in failure of cancer cell to arrest in any checkpoint making them more sensitive to genotoxic agents while normal cells with functional P53 would still be arrested and repaired. Source [10]. Figure 17](https://web.archive.org/web/20160322131841im_/http://biodiscoveryjournal.co.uk/Archive/Media/A18Figure-17_550.jpg)
In the past, researchers have identified and employed numerous inhibition strategies against DDR proteins in cancer settings and provided proof of principle for DDR inhibition-based targeted cellular sensitivity. Table 5 lists inhibitors targeting the DNA damage response pathway. It is noteworthy that many of those listed in the table are either currently in clinical trials or have been in clinical trials. For example the Chk1 inhibitors AZD7762 and PF-00477736, and the broader spectrum Chk1 and Chk2 inhibitors UCN-01 and XL-844 as well as the PARP-1 inhibitor AZD2281 have completed clinical trials whilst the DNA-PK inhibitor, CC-115, the alkyl guanine transferase inhibitor, O6 BG, and a specific Chk1 inhibitor, LY2606368, are currently recruiting patients for clinical trials.

In terms of the exploiting DDR pathway for therapeutic intervention, the inhibitors of Poly ADP-ribose polymerase-1 have made the greatest progress and many such inhibitors are currently valuated in clinical trials [228]. Following the initial discoveries of inhibitors of ATM [214, 217], the quest for finding compounds with higher specificity and to test them for their therapeutic benefit quickly followed. There are several reports that demonstrate increased cellular sensitivity towards genotoxic agents by way of modulating the ATM signalling pathway through inhibition of ATM kinase specifically.
In one of the earliest of such reports, Morgan SE et al (1997) demonstrated that over-expression of a truncated version of the functionally important leucine zipper domain of the ATM gave rise to a dominant negative ATM mutant in colon cancer cells. This group reported that such cells had an increased level of radiosensitivity and chromosomal breakage and an abrogated S-phase checkpoint. Inhibition of ATM expression, brought about by employing ATM antisense RNA and siRNA mediated gene silencing, represents another strategy adopted by several researchers. It has been demonstrated that attenuating ATM levels in human glioblastoma cells, by using antisense RNA against the kinase domain of ATM, increased their radiosensitivity independent of the status of P53 [229]. The resulting cells showed higher expression of P53 and P21 proteins, aberrant G2 checkpoint control and increased radiosensitivity after irradiation with a clinically relevant 2 Gy dose. In a similar approach the therapeutic potential of siRNA against ATM, ATR and DNA dependent protein kinase for use as radio and chemo-sensitizing agent has been assessed against human prostate cancer cells [230]. This approach reduced the respective protein levels by 90% after 48 hours post treatment, while the sensitivity of the cells to radiation was significantly increased. They also confirmed that such siRNA mediated silencing was a more potent sensitizing agent then some of the inhibitors of these kinases, e.g. wortmannin or LY294002.
In another study a negative link between EGF and ATM expression has been established and has been observed that the treatment of human lymphoblasts and fibroblasts with EGF down regulated both ATM and DNA-dependent protein kinases which, in turn, resulted in increased radiosensitization in cells with wild type ATM, while no further radiosensitization was seen in irradiated A-T cells [55]. The group further demonstrated that the reduction of ATM protein levels following EGF treatment resulted from transcriptional suppression of ATM at promoter level.
In another study the link between protein kinase C activator, 12-O-tetradecanoylphorbol 12-acetate, termed TPA, and the ability of ATM to radiosensitize the otherwise radioresistant human prostate cancer cell line and induce apoptosis has been exploited [231]. The above study reported that treatment of cells with TPA increased the levels of an apoptotic inducing regulatory enzyme ceramide synthase (CS) while caused an accompanying decrease in ATM activity, resulting in apoptosis induction after radiation treatment. This group also used antisense ATM oligonucleotides to suppress ATM expression in cancer cells. This resulted in higher levels of CS activation and apoptosis even with low radiation doses of 1Gy in radio resistant prostate cancer cells. A-T cells, which are highly chemo- and radiosensitive also have a disrupted NF-kappa B expression. Exploitation of this link between ATM and NF-kappa B pathway regulation (fig. 10) is an interesting anticancer target and experiments have been performed by using both a dominant negative form of NF kappa B inhibitor, I Kappa B alpha and inhibitors of ATM to disrupt the NF kappa B pathway to achieve chemosensitization and apoptosis of cancer cells [232].
Under genotoxic conditions of chemo or radiotherapy, some tumour cells undergo a premature senescence also called stress-induced senescence [233]. Although this permanent arrest of tumour cells is beneficial and prevents tumour metastasis, such senescent tumour cells resist apoptosis and may re-enter the cell cycle. Crescenzi et al [234] set out to identify the cellular pathways responsible for the maintenance of such drug induced tumour senescence in breast, lung and colon carcinoma cells and identified the ATM/ATR pathway to be constitutively active in these cells. They successfully used the specific ATM inhibitor KU55933 and another ATM and ATR inhibitor CGK733 [213] to block this pathway and demonstrated increased apoptosis in these cell lines. Essentially, while this treatment was cytotoxic to cancer cells, it did not cause apoptosis in the normal senescent human fibroblasts.
In another attempt to inhibit ATM, Zou J et al. [235] made use of biodegradable nanoparticles coated with ATM antisense oligonucleotides and tested this approach to inhibit ATM in cancer cells in vitro as well as in head and neck squamous cell carcinoma tumours in vivo. They reported that this approach successfully sensitised tumours to radiotherapy. Recently, Neijenhuis S et al [236] reported that lung carcinoma cells expressing an aberrant β-Polymerase, a key enzyme in base excision repair, depend on homologous recombination repair after IR treatment. In such cells, when ATM function was blocked via KU55933 treatment, an increased radiosensitivity was achieved because of failure of cells to repair through HRR. In a very recent study, Golding SE et al [237] employed the second generation ATM kinase inhibitor KU60019 in a combination treatment with Temazolamide, an alkylating agent used for treatment of brain cancer. This group demonstrated that combining this reversible inhibitor of ATM with the alkylating agent sensitized the otherwise very resistant glioma cells. Secondly this treatment was found to be cancer selective as KU60019 treatment alone did not have any growth inhibitory effects on the co-cultured human astrocytes [237].
Most recently, Roossink F et al [238] demonstrated in vitro as well as in cervical cancer patients that cancer cells that showed high levels of phosphorylated ATM (active ATM) prior to irradiation showed greater radioresistance than those with moderate levels of active ATM. Similarly, immunohistochemistry of tumour samples collected from patients with advanced stage cervical cancer also strikingly showed that tumours showing radioresistance had high levels of active ATM and this was related with poor locoreginal disease free survival and shorter disease specific survival. Furthermore, inhibition of ATM with the ATM inhibitor KU55933 strongly radiosensitised cancer cells without having deleterious effects on non-transformed epithelial cells.
6.1. Current challenges
While conceptualization of the ATM inhibition-based anticancer therapeutic approach seems straightforward, there are a number of challenges to overcome. Although, theoretically such inhibition of ATM function would disable the DDR mediated cell cycle arrest and repair and cause accumulation of DNA damage leading to cell death, there are other similar kinases with overlapping functions with ATM, such as DNA-PK and ATR. Thus, it would require a broad spectrum kinase inhibitor to inhibit all important DDR kinases. On the other hand, a more general inhibitor may have more off-target and cytotoxic effects in normal cells, which would lead to increased side effects. Secondly, ATM itself can signal to cause apoptosis via P53 dependent and independent pathways as described earlier, in which case, ATM inhibition may rather desensitise the cells against genotoxic agents and thus be counterproductive. One example of this is a recent report [10], which clearly demonstrated deference in the effects of ATM inhibition on survival of cells when exposed to a lower or higher dosage of a radiomimetic drug (context dependent difference). A further evidence of an unexpected signalling alteration caused by ATM inhibition was provided by another report recently [83]. Hence, while designing ATM inhibition based strategy for cellular sensitivity against genotoxic agents, it is imperative to consider two vital factors:
Firstly, the type of genotoxic agent employed should only trigger a DNA repair response for which ATM is a central component with no overlapping enzymes activated; hence it solely depends on ATM function. This would translate into effective sensitization specifically towards ATM inhibition after the genotoxic treatment. Secondly and equally vital is to characterise the context dependent role of ATM. As mentioned earlier, ATM is a multifunctional protein with signalling roles both in prosurvival and cytotoxic pathways. Within prosurvival pathways, ATM may function in cytoprotection mode by arresting cell cycle and initiating DNA repair, or trigger proliferative signals. On the other hand, in cytotoxic pathway, ATM may cause permanent cell cycle arrest and apoptosis or E2F1 mediated apoptosis without involving cell cycle arrest. Figure 18 illustrates the pathways involving ATM and demonstrates its multifunctional nature. Thus it becomes critical to determine experimental setting e.g. type of treatment, its dosage and time of treatment in order to employ ATM inhibition at a precise time point or at a concentration of genotoxic treatment, where ATM is functioning to promote cellular resistance towards cell death and apoptosis.

In the light of the above discussion, it has become clear that in order to identify and develop novel inhibitors of DDR pathway, there is a parallel need for identification of novel biomarkers of signalling activity of any given component in the DDR pathway. These biomarkers should be responsive to the inhibitors and should report the action of the inhibitors on the target proteins. Furthermore, in an ideal situation, these biomarkers should be characterised and validated for reporting the most vulnerable cellular states in terms of the therapy and inhibition, which could be dictated by the redox status of the cell, phase of the cell cycle or metabolic activity. This feature of DDR manipulation strategy is further elaborated in the next section.
6.2. Future direction
While increasing understanding of the aetiology of cancer has served to answer many questions, it has also generated numerous new ones. Characterisation of multifunctional signalling molecules has made it quite clear that each node in a network of signalling pathway represents several pathway choices that could generate or contribute in overlapping or often different and context dependent cellular response to a drug. This is complicated by the fact that owing to a complex networking nature of cellular signals, the complement of genetic mutations within a tumour may influence pathway preferences of multifunctional proteins in a given treatment protocol.
The future research endeavours should focus on elucidating the state of key decision makers in this intricate network of signalling molecules in different cancers. Such research attempts would not only shed light on the multifunctional molecules inducing pathway preferences that would explain altered cellular responses of cancer and normal cells to the same treatment regimes, but would also allow for the manipulation of such network linkages and decision making nodes in the pathway. We believe that a prerequisite to get to such level of molecular detailing is a generation of novel biosensors and detection techniques that are highly sensitive and allow for high throughput mode of detection of both behaviour of individual pathway components as well as overall physiological changes within the cell. There are numerous examples of biosensors and in situ DNA damage detection methods in the literature surrounding different signalling component of the DDR pathway. Key recent biosensors surrounding ATM and most prominent DNA damage detection techniques are summarised in table 6. With regard to drug development, of particular interests are biosensors, which can monitor activity of key target enzymes such as kinases in real time in live cells. Since phosphorylation often modulate stability of protein substrates [239] genetically engineered cells expressing luciferase-conjugated kinase substrates can be utilised as biomarkers and biosensors to monitor activity of kinase inhibitors in live cells [240].

The new generation of biosensors should not only report the activity of a molecule, for example kinase activity of ATM during a treatment regime, but should also inform us of the pathway preference of ATM once activated or report a change whenever it switches its multifunctional nature of signalling between, for example, cell cycle arrest or proliferation, DNA repair or apoptotic induction, ROS generating pathway or ROS scavenging (see figures 16 and 18). This would not only allow to monitor enzymatic activity of critical enzymes in DDR, but would also report in real time how and when pathway choices are made by enzymes implicated in dual or multiple functions. However, a mere reporter of an enzymatic induction or inhibition or a biosensor of DNA damage (e.g. those listed in table 6) fails to serve the above mentioned critical detailing.
A way to create such biosensors is to design an assay that reports changes in a signalling branch rather than a signalling component i.e. an assay that is based on the activation of a signalling link emerging from a single molecule rather than a mere activation or inhibition of that molecule. This could be done in the form of reporting real time protein-protein interaction that necessitates a cellular response or senses the formation of a complex of proteins that is a commitment to only a specific cellular outcome. This is based on the principle that multifunctional proteins contribute in multiple pathway signalling by altering their substrate preferences depending upon the state of cell health. Additionally, these biosensors should also incorporate information from protein trafficking and localisation changes. There are numerous proteins whose downstream signalling preference depends upon their cellular localisation at a given time. Hence tightly coupled to induction and inhibition is anterograde and retrograde transport that would lead to either sequestration or complex formation for an enzyme with its substrate. Recently, in terms of ATM signalling, it was found that it undergoes DNA damage induced Golgi to nuclear transport. This trafficking event led to nuclear accumulation of pATM and pP53 Ser 15 [99]. This change in ATM localisation (upon activation of its DDR function) may have consequences for cytoplasmic component of its signalling pathway (implicated in proliferation and ROS generation), e.g. NFkB signalling and in mTORC regulation [193]. Hence, a biosensor that reports these changes in addition to induction or inhibition will be more predictive of the cellular response upon drug intervention.
Such biosensors once designed and validated could then be used to carry out signal profiling of premalignant and malignant tumours isolated from patients. This could be done by exposing them to test treatment protocols and determine the behaviour of multifunctional proteins e.g. ATM, that may differ greatly both in induction kinetics as well as pathway choices on the basis of the complement of genetic mutations in the tumour. Based on data from the biosensor, post treatment predictions of signalling behaviour and ultimate cellular response to therapy could be made, protocols could be fine-tuned to a therapeutic window and optimal treatment regimens devised for the actual therapy with lowest cytotoxic effects on normal tissues.
References
- Chicheva Z, Chelenkova P, Petkova R, Chakarov S. Children of the sun, children of the moon – a mini-panel for assessment of inter-individual variation between the capacity of healthy individuals to repair everyday genotoxic insults. Biotechnol Biotech Eq 2012; 26(4): 3142-3147.
- Tummala H, Khalil HS, Zhelev N. Repair, abort, ignore? Strategies for dealing with UV damage. Biotechnol Biotech Eq 2011; 25(3): 2443-2446.
REFERENCE LINK - Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001; 27(3): 247-254.
REFERENCE LINK - Shen Z, Nickoloff JA. Mammalian homologous recombination repair and cancer intervention. In: DNA repair, genetic instability, and cancer. Wei Q, Li L, Chen DJ, Eds. World scientific publishing Co, Singapore: 2007, pp 119-156.
REFERENCE LINK - Valerie K, Povirk LF. Regulation and mechanisms of mammalian double- trand break repair. Oncogene 2003; 22(37): 5792-5812.
REFERENCE LINK - Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell Mol Life Sci 2002; 59(2): 373-385.
REFERENCE LINK - Chakarov S, Roeva I, Russev G. An experimental model for assessment of global DNA repair capacity. Biotechnol Biotech Eq 2011; 25(3): 2505-2507.
REFERENCE LINK - Chakarov S, Russev G. DNA repair and differentiation – does getting older means getting wiser as well? Biotechnol Biotech Eq 2010; 24(2): 1804-1806.
REFERENCE LINK - Chakarov, S, Stoilov, P, Alexandrov, A, Russev G. Repair pattern in the beta-globin gene cluster of human fibroblasts after ultraviolet irradiation. Eur J Biochem 1997; 248(3): 669-675.
REFERENCE LINK - Khalil HS, Tummala H, Chakarov S, Zhelev N, Lane DP. Targeting ATM pathway for therapeutic intervention in cancer. Biodiscovery 2012; 1: 3.
REFERENCE LINK - Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001; 412(6847): 607-614.
REFERENCE LINK - Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res 1999; 27(24): 4679-4686.
REFERENCE LINK - Smith GC, d’Adda di Fagagna F, Lakin ND, Jackson SP. Cleavage and inactivation of ATM during apoptosis. Mol Cell Biol 1999; 19(9): 6076-6084.
- Karmakar P, Piotrowski J, Brosh RM Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem 2002; 277(21): 18291-18302.
REFERENCE LINK - Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002; 108(6): 781-794.
REFERENCE LINK - Broderick S, Rehmet K, Concannon C, Nasheuer HP. Eukaryotic single- stranded DNA binding proteins: central factors in genome stability. Subcell Biochem 2010; 50: 143-163.
REFERENCE LINK - Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol 2002; 22(14): 5194-5202.
REFERENCE LINK - Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem 2000; 275(34): 26196-26205.
REFERENCE LINK - Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 1996; 271(15): 8936-8941.
REFERENCE LINK - Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 1998; 17(18): 5497-5508.
REFERENCE LINK - Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 1998; 93(3): 477-486.
REFERENCE LINK - Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res 2008; 18(1):134-147.
REFERENCE LINK - Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH et al. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 2000; 404(6778): 613-617.
REFERENCE LINK - Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 2007; 85(4): 509-520.
REFERENCE LINK - Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res 1998; 26(23): 5388-5393.
REFERENCE LINK - Swagemakers SM, Essers J, de Wit J, Hoeijmakers JH, Kanaar R. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J Biol Chem 1998; 273(43): 28292-28297.
REFERENCE LINK - Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 1999; 398(6729):728-731.
REFERENCE LINK - Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002; 297(5588): 1837-1848.
REFERENCE LINK - Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM. The structure of the Holliday junction, and its resolution. Cell 1988; 55(1): 79-89.
REFERENCE LINK - Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J 2000; 19(20): 5552-5561.
REFERENCE LINK - Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science 1989; 246: 629-634.
REFERENCE LINK - Kastan MB, Bartek, J. Cell cycle checkpoints and cancer. Nature 2004; 432: 316-323.
REFERENCE LINK - Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995; 268:1749-1753.
REFERENCE LINK - Lee Y, McKinnon PJ. ATM dependent apoptosis in the nervous system. Apoptosis 2000; 5: 523-529.
REFERENCE LINK - Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K et al. ATM deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 1998; 125(20): 4007-4017.
- Gatti RA, Berkel I, Boder E, Braedt G, Charmley P, Concannon P et al. Localization of an ataxia- elangiectasia gene to chromosome 11q22-23. Nature 1988; 336(6199): 577-580.
REFERENCE LINK - Chen X, Yang L, Udar N, Liang T, Uhrhammer N, Xu S, et al. CAND3: a ubiquitously expressed gene immediately adjacent and in opposite transcriptional orientation to the ATM gene at 11q23.1. Mamm Genome 1997; 8(2): 129-133.
REFERENCE LINK - Uziel T, Savitsky K, Platzer M, Ziv Y, Helbitz T, Nehls M et al. Genomic Organization of the ATM gene. Genomics 1996; 33(2): 317-320.
REFERENCE LINK - Prokopcova J, Kleibl Z, Banwell CM, Pohlreich P. The role of ATM in breast cancer development. Breast Cancer Res Treat 2007; 104(2): 121-128.
REFERENCE LINK - Platzer M, Rotman G, Bauer D, Uziel T, Savitsky K, Bar-Shira A et al. Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res 1997; 7(6): 592-605.
- Savitsky K, Platzer M, Uziel T, Gilad S, Sartiel A, Rosenthal A et al. Ataxia-telangiectasia: structural diversity of untranslated sequences suggests complex post-transcriptional regulation of ATM gene expression. Nucleic Acids Res 1997; 25(9): 1678-1684.
REFERENCE LINK - Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I et al. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 1997; 15(2): 159-167.
REFERENCE LINK - Scott SP, Zhang N, Khanna KK, Khromykh A, Hobson K, Watters D et al. Cloning and expression of the ataxia-telangiectasia gene in baculovirus. Biochem Bioph Res Co 1998; 245(1): 144-148.
REFERENCE LINK - Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci U S A 1996; 93(23): 13084-13089.
REFERENCE LINK - Barlow C, Ribaut-Barassin C, Zwingman TA, Pope AJ, Brown KD, Owens JW et al. ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc Natl Acad Sci USA 2000; 97(2): 871-876.
REFERENCE LINK - Imai T, Yamauchi M, Seki N, Sugawara T, Saito T, Matsuda Y et al.. Identification and characterization of a new gene physically linked to the ATM gene. Genome Res 1996; 6(5): 439-447.
REFERENCE LINK - Imai T, Sugawara T, Nishiyama A, Shimada R, Ohki R, Seki N et al. The structure and organization of the human NPAT gene. Genomics 1997; 42(3): 388-392.
REFERENCE LINK - Byrd PJ, Cooper PR, Stankovic T, Kullar HS, Watts GD, Robinson PJ et al. A gene transcribed from the bidirectional ATM promoter coding for a serine rich protein: amino acid sequence, structure and expression studies. Hum Mol Genet 1996; 5(11): 1785-1791.
REFERENCE LINK - Gentilini F, Turba ME, Forni M, Cinotti S. Complete sequencing of full-length canine ataxia telangiectasia mutated mRNA and characterization of its putative promoter. Vet Immunol Immunopathol 2009; 128(4): 437-440.
REFERENCE LINK - Platzer M, Rotman G, Bauer D, Uziel T, Savitsky K, Bar-Shira A et al. Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res 1997; 7(6): 592-605.
- Khalil HS, Tummala H, Oluwaseun OA, Zhelev N. Novel insights of Ataxia Telangiectasia Mutated (ATM) regulation and its potential as a target for therapeutic intervention in cancer. Curr Opin Biotechnol 2011; 22(1) S115-S116.
REFERENCE LINK - Craig AL, Holcakova J, Finlan LE, Nekulova M, Hrstka R, Gueven N et al. DeltaNp63 transcriptionally regulates ATM to control p53 Serine-15 phosphorylation. Mol Cancer 2010, 9:195.
REFERENCE LINK - Gueven N, Fukao T, Luff J, Paterson C, Kay G, Kondo N et al. Regulation of the Atm promoter in vivo. Genes Chromosomes Cancer 2006; 45: 61-71.
REFERENCE LINK - Gueven N, Keating K, Fukao T, Loeffler H, Kondo N, Rodemann HP, Lavin MF. Site-directed mutagenesis of the ATM promoter: consequences for response to proliferation and ionizing radiation. Genes Chromosome Canc 2003; 38:157-167.
REFERENCE LINK - Gueven N, Keating KE, Chen P, Fukao T, Khanna KK, Watters D et al. Epidermal growth factor sensitizes cells to ionizing radiation by down-regulating protein mutated in ataxia-telangiectasia. J Biol Chem 2001; 276(12): 8884-8891.
REFERENCE LINK - Hirai Y, Hayashi T, Kubo Y, Hoki Y, Arita I, Tatsumi K et al. X-irradiation induces up-regulation of ATM gene expression in wild-type lymphoblastoid cell lines, but not in their heterozygous or homozygous ataxia-telangiectasia counterparts. Jpn J Cancer Res 2001; 92(6): 710-717.
REFERENCE LINK - Rogatcheva MB, Fritz KL, Rund LA, Pollock CB, Beever JE, Counter CM et al. Characterization of the porcine ATM gene: towards the generation of a novel non-murine animal model for Ataxia-Telangiectasia. Gene 2007; 405(1-2): 27-35.
REFERENCE LINK - Fang ZM, Lee CS, Sarris M, Kearsley JH, Murrell D, Lavin MF et al. Rapid radiation-induction of ATM protein levels in situ. Pathology 2001; 33(1): 30-36.
- Chen G, Lee EYHP. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J Biol Chem 1996; 271(52): 33693-33697.
REFERENCE LINK - Morgan SE, Lovly C, Pandita TK, Shiloh Y, Kastan MB.. Fragments of ATM which have dominant-negative or complementing activity. Mol Cell Biol 1997; 17(4): 2020-2029.
- Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature 1997; 387(6632): 520-523.
REFERENCE LINK - Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421(6922): 499-506.
REFERENCE LINK - Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J 2006; 25(15): 3504-3514.
REFERENCE LINK - Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 2005; 102(37): 13182-13187.
REFERENCE LINK - Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 2007 27(24): 8502-8509.
REFERENCE LINK - Kozlov SV, Graham ME, Jakob B, Tobias F, Kijas AW, Tanuji M et al. Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem 2011; 286(11): 9107-9119.
REFERENCE LINK - Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316(5828): 1160-1166.
REFERENCE LINK - Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Körner R et al. kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 2008; 31(3): 438-448.
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Kéri G et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 2009; 8(7): 1751-1764.
- Gately DP, Hittle JC, Chan GK, Yen TJ. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol Biol Cell 1998; 9: 2361-2374.
- Chen G, Lee EYHP. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J Biol Chem 1996; 271(52): 33693-33697.
REFERENCE LINK - Soares HD, Morgan JI, McKinnon PJ. ATM expression patterns suggest a contribution from the peripheral nervous system to the phenotype of ataxia-telangiectasia. Neuroscience 1998; 86(4): 1045-1054.
REFERENCE LINK - Qi J, Shackelford R, Manuszak R, Cheng D, Smith M, Link CJ et al. Functional expression of ATM gene carried by HSV amplicon vector in vitro and in vivo. Gene Ther 2004; 11(1): 25-33.
REFERENCE LINK - Meyn MS. Ataxia-telangiectasia, cancer and the pathobiology of the ATM gene. Clin Genet 1999; 55(5): 289-304.
REFERENCE LINK - Khalil HS, Tummala H, Zhelev N. Differences in the DDR enzymes activation kinetics between normal and cancer cells could be utilized to achieve targeted cellular sensitivity towards genotoxic agents. Cancer Res 2012; 72 (8 Supplement): 3103.
REFERENCE LINK - Levitt NC, Hickson ID. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol Med 2002; 8(4): 179-186.
REFERENCE LINK - Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science 2010; 330(6003): 517-521.
REFERENCE LINK - Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle 2010; 9(24): 4805-4811.
REFERENCE LINK - Brown KD, Ziv Y, Sadanandan SN, Chessa L, Collins FS, Shiloh Y et al. The ataxia-telangiectasia gene product, a constitutively expressed nuclear protein that is not up-regulated following genome damage. Proc Natl Acad Sci USA 1997; 94(5): 1840-1845.
REFERENCE LINK - Watters D, Khanna KK, Beamish H, Birrell G, Spring K, Kedar P et al. Cellular localisation of the ataxia-telangiectasia (ATM) gene product and discrimination between mutated and normal forms. Oncogene 1997; 14(16): 1911-1921.
REFERENCE LINK - Fukao T, Kaneko H, Birrell G, Gatei M, Tashita H, Yoshida T et al. ATM is upregulated during the mitogenic response in peripheral blood mononuclear cells. Blood 1999; 94:1998-2006.
- Moiola C, De Luca P, Cotignola J, Gardner K, Vazquez E, De Siervi A. Dynamic coregulatory complex containing BRCA1, E2F1 and CtIP controls ATM transcription. Cell Physiol Biochem 2012; 30(3): 596-608.
REFERENCE LINK - Khalil HS, Tummala H, Hupp TR, Zhelev N. Pharmacological inhibition of ATM by KU55933 stimulates ATM transcription. Exp Biol Med (Maywood) 2012; 237: 622-634.
REFERENCE LINK - Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998; 281(5383): 1674-1677.
REFERENCE LINK - Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 2000; 14: 278-288,
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001; 15: 1067-1077.
REFERENCE LINK - Lim DS, Kirsch DG, Canman CE, Ahn JH, Ziv Y, Newman LS et al. ATM binds to beta-adaptin in cytoplasmic vesicles. Proc Natl Acad Sci USA 1998; 95(17): 10146-10151.
REFERENCE LINK - Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 2002; 109:459-472.
REFERENCE LINK - Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev 2002; 16:560-570.
REFERENCE LINK - Xu B, O’Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res 2002; 62:4588-4591.
- Xu B, Kim St, Kastan MB. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol Cell Biol 2001; 21:3445-3450.
REFERENCE LINK - Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A et al. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 2001; 411: 969-974.
REFERENCE LINK - Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev 1996; 10: 2401-2410.
REFERENCE LINK - Westphal CH, Rowan S, Schmaltz C, Elson A, Fisher DE, Leder P. ATM and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat Genet 1997; 16: 397-401.
REFERENCE LINK - Chong MJ, Murray MR, Gosink EC, Russell HR, Srinivasan A, Kapsetaki M et al. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc Natl Acad Sci USA 2000; 97: 889-894.
REFERENCE LINK - Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 2004; 18: 3041-3054.
REFERENCE LINK - Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res 2004; 2: 203-214.
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001; 276(45): 42462-42467.
REFERENCE LINK - Khalil HS, Tummala H, Caris LD, Zhelev N. Phosphorylated ATM at S-1981 (pATM) undergoes COPI mediated Golgi export upon DNA damage. FEBS J 2012; 279(S1): 149.
- Oluwaseun OA, Khalil HS. War without weapons – constitution of healthy and pathological phenotypes associated with polymorphisms in genes involved in the maintenance of genome integrity. Biotechnol Biotech Eq 2012; 26(4): 3073-3078.
- Wang H, Guan J, Wang H, Perrault AR, Wang Y, Iliakis G. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res 2001; 61(23): 8554-8563.
- Gatei M, Zhou BB, Hobson K, Scott S, Young D, Khanna KK. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J Biol Chem 2001 276(20): 17276-17280.
REFERENCE LINK - Foray N, Marot D, Gabriel A, Randrianarison V, Carr AM, Perricaudet M et al. A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. EMBO J 2003; 22(11): 2860-2871.
REFERENCE LINK - Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 2001; 410: 842–847.
- Gatei M, Sloper K, Sorensen C, Syljuäsen R, Falck J, Hobson K et al. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem 2003; 278(17): 14806-14811.
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 2000; 14(3): 289-300. Erratum in: Genes Dev 2000; 14(6): 750.
- Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson CJ, Nigg EA et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol 2004; 6(9): 884-891.
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 1997; 277(5331): 1497-501.
REFERENCE LINK - Carcagno AL, Ogara MF, Sonzogni SV, Marazita MC, Sirkin PF, Ceruti JM, Cánepa ET. E2F1 transcription is induced by genotoxic stress through ATM/ATR activation. IUBMB Life 2009; 61(5): 537-543.
REFERENCE LINK - Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet 2000; 26(4): 464-469.
- Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol 2003; 5(5): 401-409.
REFERENCE LINK - Sahu RP, Batra S, Srivastava SK. Activation of ATM/Chk1 by curcumin causes cell cycle arrest and apoptosis in human pancreatic cancer cells. Br J Cancer 2009; 100(9): 1425-1433.
REFERENCE LINK - Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000; 19(49): 5643-5650.
REFERENCE LINK - Ahmed KM, Li JJ. ATM-NF-kappaB connection as a target for tumor radiosensitization. Curr Cancer Drug Targets 2007; 7(4): 335-342.
REFERENCE LINK - Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000; 10(15): 886-895.
REFERENCE LINK - Ivanova, I.A., Vespa, A., Dagnino, L. A novel mechanism of E2F1 regulation via nucleocytoplasmic shuttling: determinants of nuclear import and export. Cell Cycle 2007; 6: 2186-2195.
REFERENCE LINK - Rodriguez JA, Schüchner S, Au WW, Fabbro M, Henderson BR. Nuclear-Cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene 2004; 23(10): 1809-1820.
REFERENCE LINK - Liang SH, Clarke MF. Regulation of P53 localization. Eur J Biochem 2001; 268(10): 2779-2783.
REFERENCE LINK - Chakarov S, Petkova R, Russev, G Ch. P53-Gardian angel and archangel. Biotechnol Biotech Eq 2012; 26(1): 2695-2702.
REFERENCE LINK - Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 2005; 309(5741): 1732-1735.
REFERENCE LINK - Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res 2005; 65(9): 3745-3750.
REFERENCE LINK - Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 2000; 275(21): 16202-16212.
REFERENCE LINK - Frossi B, Tell G, Spessotto P, Colombatti A, Vitale G, Pucillo C. H(2)O(2) induces translocation of APE/Ref-1 to mitochondria in the Raji B-cell line. J Cell Physiol 2002; 193(2): 180-186.
REFERENCE LINK - Smaili SS, Hsu YT, Sanders KM, Russell JT, Youle RJ. Bax translocation to mitochondria subsequent to a rapid loss of mitochondrial membrane potential. Cell Death Differ 2001; 8(9): 909-920.
REFERENCE LINK - Okada S, Ouchi T. Cell cycle differences in DNA damage-induced BRCA1 phosphorylation affect its subcellular localisation. J Biol Chem 2003; 278(3): 2015-2020.
REFERENCE LINK - Enomoto M, Goto H, Tomono Y, Kasahara K, Tsujimura K, Kiyono T et al. Novel positive feedback loop between Cdk1 and Chk1 in the nucleus during G2/M transition. J Biol Chem 2009; 284(49): 34223-34230.
REFERENCE LINK - Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localisation of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 1999; 397(6715): 172-175.
REFERENCE LINK - Oberkovitz G, Regev L, Gross A. Nucleocytoplasmic shuttling of BID is involved in regulating its activities in the DNA-damage response. Cell Death Differ 2007; 14(9): 1628-1634.
REFERENCE LINK - Li C, Chen L, Chen J. DNA damage induces MDMX nuclear translocation by P53-dependent and -independent mechanisms. Mol Cell Biol 2002; 22(21): 7562-7571.
REFERENCE LINK - De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem 2006; 281(23): 16147-16156.
REFERENCE LINK - Carter T, Vancurová I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol 1990; 10(12): 6460-6471.
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 2000; 14(23): 2989-3002.
REFERENCE LINK - Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates P53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 2004; 6(7): 665-672.
REFERENCE LINK - Gjerset RA, Bandyopadhyay K. Regulation of p14ARF through subnuclear compartmentalization. Cell Cycle 2006; 5(7): 686-690.
REFERENCE LINK - Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 2006; 311(5764): 1141-1146.
REFERENCE LINK - Ismail F, Ikram M, Purdie K, Harwood C, Leigh I, Storey A. Cutaneous squamous cell carcinoma (SCC) and the DNA damage response: pATM expression patterns in pre-malignant and malignant keratinocyte skin lesions. PLoS One 2011; 6(7): e21271.
REFERENCE LINK - Li J, Han YR, Plummer MR, Herrup K. Cytoplasmic ATM in neurons modulates synaptic function. Curr Biol 2009; 19(24): 2091-2096.
REFERENCE LINK - Kuljis RO, Chen G, Lee EY, Aguila MC, Xu Y. ATM immunolocalisation in mouse neuronal endosomes: implications for ataxia-telangiectasia. Brain Res 1999; 842(2): 351-358.
REFERENCE LINK - Young DB, Jonnalagadda J, Gatei M, Jans DA, Meyn S, Khanna KK. Identification of domains of ataxia-telangiectasia mutated required for nuclear localisation and chromatin association. J Biol Chem 2005; 280(30): 27587-27594.
REFERENCE LINK - Zhang L, Tie Y, Tian C, Xing G, Song Y, Zhu Y et al. CKIP-1 recruits nuclear ATM partially to the plasma membrane through interaction with ATM. Cell Signal 2006; 18(9): 1386-1395.
REFERENCE LINK - Oka A, Takashima S. Expression of the ataxia-telangiectasia gene (ATM) product in human cerebellar neurons during development. Neurosci Lett 1998; 252(3): 195-198.
REFERENCE LINK - Watters D, Kedar P, Spring K, Bjorkman J, Chen P, Gatei M et al. Localization of a portion of extranuclear ATM to peroxisomes. J Biol Chem 1999; 274(48): 34277-34282.
REFERENCE LINK - Swift M. Genetics and epidemiology of ataxia-telangiectasia. Kroc Found Ser 1985; 19: 133-146.
- Perlman S, Becker-Catania S, Gatti RA. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol 2003; 10(3): 173-182.
REFERENCE LINK - Swift M. Genetic aspects of ataxia-telangiectasia. Immunodefic Rev 1990; 2(1): 67-81.
- Gilad S, Bar-Shira A, Harnik R, Shkedy D, Ziv Y, Khosravi R et al.. Ataxia-telangiectasia: founder effect among North African Jews. Hum Mol Genet 1996; 5(12): 2033-2037.
REFERENCE LINK - Su Y, Swift M. Mortality rates among carriers of ataxia-telangiectasia mutant alleles. Ann Intern Med 2000; 133(10): 770-778.
- Foray N, Priestley A, Alsbeih G, Badie C, Capulas EP, Arlett CF et all. Hypersensitivity of ataxia telangiectasia fibroblasts to ionizing radiation is associated with a repair deficiency of DNA double-strand breaks. Int J Radiat Biol 1997; 72: 271-283.
REFERENCE LINK - Peterson RD, Funkhouser JD, Tuck-Muller CM, Gatti RA. Cancer susceptibility in ataxia-telangiectasia. Leukemia 1992; 6 (1 Suppl): 8-13.
- Reed WB, Epstein WL, Boder E, Sedgwick R. Cutaneous manifestations of ataxia-telangiectasia. JAMA 1966; 195(9): 746-753.
REFERENCE LINK - Mitui M, Nahas SA, Du LT, Yang Z, Lai CH, Nakamura K et al. Functional and computational assessment of missense variants in the ataxia-telangiectasia mutated (ATM) gene: mutations with increased cancer risk. Hum Mutat 2009; 30(1): 12-21.
REFERENCE LINK - Frederick H., Barbara K. H. Cancer in Ataxia-telangiectasia patients. Cancer Genet and Cytogen 1990; 46(1): 9-19.
REFERENCE LINK - Taylor AM. Ataxia telangiectasia genes and predisposition to leukaemia, lymphoma and breast cancer. Br J Cancer 1992; 66(1): 5-9.
REFERENCE LINK - Gao Y, Hayes RB, Huang WY, Caporaso NE, Burdette L, Yeager M et al. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis 2011; 32(6): 882-887.
REFERENCE LINK - McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep 2004; 5(8): 772-776.
REFERENCE LINK - Clyde RG, Bown JL, Hupp TR, Zhelev N, Crawford JW. The role of modelling in identifying drug targets for diseases of the cell cycle. J R Soc Interface 2006; 3(10): 617-627.
REFERENCE LINK - Idowu MA, Goltsov A, Khalil HS, Hemanth T, Zhelev N, Bown J. Cancer research and personalised medicine: a new approach to modelling time-series data using analytical methods and Half systems. Curr Opin Biotechnol 2011; 22(S1): 59.
REFERENCE LINK - Clyde R, Tummala H, Khalil HS, Goszcz K, Lucka I, Tupone MG et al. A novel quantitative systems biology approach to cancer research and treatment. Curr Opin Biotechnol 2011; 22(S1): 58.
REFERENCE LINK - Tummala H, Khalil HS, Goszcz K, Tupone MG, Stoyanova V, Nikolova E et al. A quantitative integrated systems biology approach for modeling cell cycle pathways in normal and tumor cells. Cancer Res 2012; 72(8 Supplement): 4913.
REFERENCE LINK - Idowu MA, Bown J, Zhelev N. A new method for identifying a data-consistent self-reconfigurable predictive bio-network model of the cell cycle based on time series data and its application in cancer systems biology. Cancer Res 2012; 72(8 Supplement): 4921.
REFERENCE LINK - Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol 2003; 15(2): 221-231.
REFERENCE LINK - Zhou GP, Deng MH. An extension of Chou’s graphic rules for deriving enzyme kinetic equations to systems involving parallel reaction pathways. Biochem J 1984; 222(1): 169-176.
- Xiao X, Shao S, Ding Y, Huang Z, Chen X, Chou KC. An application of gene comparative image for predicting the effect on replication ratio by HBV virus gene missense mutation. J Theor Biol 2005; 235(4): 555-565.
REFERENCE LINK - Wang M, Yao JS, Huang ZD, Xu ZJ, Liu GP, Zhao HY et al. A new nucleotide-composition based fingerprint of SARS-CoV with visualization analysis. Med Chem 2005; 1(1): 39-47.
REFERENCE LINK - Chickarmane V, Ray A, Sauro HM, Nadim A. A Model for p53 Dynamics Triggered by DNA Damage. Siam J Appl Dyn Syst 2007; 6: 61-78.
REFERENCE LINK - Qi JP, Shao SH, Xie J, Zhu Y. A mathematical model of P53 gene regulatory networks under radiotherapy. Biosystems 2007; 90(3): 698-706.
REFERENCE LINK - Sun T, Yang W, Liu J, Shen P. Modeling the basal dynamics of p53 system. PLoS One 2011; 6(11): e27882.
REFERENCE LINK - Lupi M, Matera G, Natoli C, Colombo V, Ubezio P. The contribution of p53 in the dynamics of cell cycle response to DNA damage interpreted by a mathematical model. Cell Cycle 2007; 6(8): 943-950.
REFERENCE LINK - Lahav G. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci STKE 2004; 2004(264): pe55.
REFERENCE LINK - Sun T, Chen C, Wu Y, Zhang S, Cui J, Shen P. Modeling the role of p53 pulses in DNA damage- induced cell death decision. BMC Bioinformatics 2009; 10: 190.
REFERENCE LINK - Zhang XP, Liu F, Cheng Z, Wang W. Cell fate decision mediated by p53 pulses. Proc Natl Acad Sci USA 2009; 106(30): 12245-12250.
REFERENCE LINK - Iwamoto K, Hamada H, Eguchi Y, Okamoto M. Mathematical modeling of cell cycle regulation in response to DNA damage: exploring mechanisms of cell-fate determination. Biosystems 2011; 103(3): 384-391.
REFERENCE LINK - Tashima Y, Hamada H, Okamoto M, Hanai T. Prediction of key factor controlling G1/S phase in the mammalian cell cycle using system analysis. J Biosci Bioeng 2008; 106(4): 368-374.
REFERENCE LINK - Iwamoto K, Tashima Y, Hamada H, Eguchi Y, Okamoto M. Mathematical modeling and sensitivity analysis of G1/S phase in the cell cycle including the DNA-damage signal transduction pathway. Biosystems 2008; 94(1-2): 109-17.
REFERENCE LINK - Powathil GG, Gordon KE, Hill LA, Chaplain MA. Modelling the effects of cell-cycle heterogeneity on the response of a solid tumour to chemotherapy: biological insights from a hybrid multiscale cellular automaton model. J Theor Biol 2012; 308: 1-19.
REFERENCE LINK - Kitano H. Computational systems biology. Nature 2002; 420(6912): 206-210.
REFERENCE LINK - Faratian D, Goltsov A, Lebedeva G, Sorokin A, Moodie S, Mullen P et al. Systems biology reveals new strategies for personalizing cancer medicine and confirms the role of PTEN in resistance to trastuzumab. Cancer Res 2009 69(16): 6713-6720.
REFERENCE LINK - Lebedeva G, Sorokin A, Faratian D, Mullen P, Goltsov A, Langdon SP et al. Model-based global sensitivity analysis as applied to identification of anti-cancer drug targets and biomarkers of drug resistance in the ErbB2/3 network. Eur J Pharm Sci 2012; 46(4): 244-258.
REFERENCE LINK - Khalil HS, Petkova R, Zhelev N. Differential genetic advantage in youth and in aging or how to die healthy. Biotechnol Biotech Eq 2011; 26(1): 2703-2711.
REFERENCE LINK - Barzilai A, Rotman G, Shiloh Y. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair (Amst) 2002; 1(1): 3-25.
REFERENCE LINK - Meredith MJ, Dodson ML. Imparied glutathione biosynthesis in cultured human ataxia-telangiectasia cells. Cancer Res 1987; 47(17): 4576-4581.
- Yi M, Rosin MP, Anderson CK. Response of fibroblast cultures from ataxia-telangiectasia patients to oxidative stress. Cancer Lett 1990; 54(1-2): 43-50.
REFERENCE LINK - Ward AJ, Olive PL, Burr AH, Rosin MP. Response of fibroblast cultures from ataxia-telangiectasia patients to reactive oxygen species generated during inflammatory reactions. Environ Mol Mutagen 1994; 24(2): 103-111.
REFERENCE LINK - Reichenbach J, Schubert R, Schwan C, Müller K, Böhles HJ, Zielen S. Anti-oxidative capacity in patients with ataxia telangiectasia. Clin Exp Immunol 1999;117(3): 535-539.
REFERENCE LINK - Barlow C, Dennery PA, Shigenaga MK, Smith MA, Morrow JD, Roberts LJ 2nd et al. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc Natl Acad Sci USA 1999; 96(17): 9915-9919.
REFERENCE LINK - Takao N, Li Y, Yamamoto K. Protective roles for ATM in cellular response to oxidative stress. FEBS Lett 2000; 472(1): 133-136.
REFERENCE LINK - Kamsler A, Daily D, Hochman A, Stern N, Shiloh Y, Rotman G et al. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice. Cancer Res 2001; 61(5): 1849-1854.
- Quick KL, Dugan LL. Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann Neurol 2001 49(5): 627-635.
REFERENCE LINK - Shackelford RE, Innes CL, Sieber SO, Heinloth AN, Leadon SA, Paules RS. The Ataxia telangiectasia gene product is required for oxidative stress-induced G1 and G2 checkpoint function in human fibroblasts. J Biol Chem 2001; 276(24): 21951-21959.
REFERENCE LINK - Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 2004; 431(7011): 997-1002.
REFERENCE LINK - Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem 2004; 279(51): 53272-53281.
REFERENCE LINK - Kim J, Wong PK. Oxidative stress is linked to ERK1/2-p16 signaling-mediated growth defect in ATM-deficient astrocytes. J Biol Chem 2009 284(21): 14396-14404.
REFERENCE LINK - Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA 2010; 107(9): 4153-4258. Erratum in: Proc Natl Acad Sci USA 2012; 109(21): 8352.
REFERENCE LINK - Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009; 196(1): 65-80.
REFERENCE LINK - Viniegra JG, Martínez N, Modirassari P, Hernández Losa J, Parada Cobo C, Sánchez-Arévalo Lobo VJ et al. Full activation of PKB/Akt in response to insulin or ionizing radiation is mediated through ATM. J Biol Chem 2005 280(6): 4029-4036.
- Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J 2011; 30(3): 546-555.
REFERENCE LINK - Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell 2010; 40(4): 509-520.
REFERENCE LINK - Fu X, Wan S, Lyu YL, Liu LF, Qi H. Etoposide induces ATM-dependent mitochondrial biogenesis through AMPK activation. PLoS One 2008; 3(4): e2009.
- Cheema AK, Timofeeva O, Varghese R, Dimtchev A, Shiekh K, Shulaev V et al. Integrated analysis of ATM mediated gene and protein expression impacting cellular metabolism. J Proteome Res 2011; 10(5): 2651-2657.
REFERENCE LINK - Wood LM, Sankar S, Reed RE, Haas AL, Liu LF, McKinnon P et al. A novel role for ATM in regulating proteasome-mediated protein degradation through suppression of the ISG15 conjugation pathway. PLoS One 2011; 6(1): e16422.
REFERENCE LINK - Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci 2012; 37(1): 15-22.
REFERENCE LINK - Bar RS, Levis WR, Rechler MM, Harrison LC, Siebert C, Podskalny J et al. Extreme insulin resistance in ataxia telangiectasia: defect in affinity of insulin receptors. N Engl J Med 1978; 298(21): 1164-1171.
REFERENCE LINK - Peretz S, Jensen R, Baserga R, Glazer PM. ATM-dependent expression of the insulin-like growth factor-I receptor in a pathway regulating radiation response. Proc Natl Acad Sci USA 2001; 98(4): 1676-1681.
REFERENCE LINK - Armata HL, Golebiowski D, Jung DY, Ko HJ, Kim JK, Sluss HK. Requirement of the ATM/p53 tumor suppressor pathway for glucose homeostasis. Mol Cell Biol 2010; 30(24): 5787-5794.
REFERENCE LINK - Miyamoto S. Nuclear initiated NF-κB signaling: NEMO and ATM take center stage. Cell Res 2011; 21(1): 116-130.
REFERENCE LINK - Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Ogura T, Lavin MF et al. IGF-1 phosphorylates AMPK-alpha subunit in ATM-dependent and LKB1-independent manner. Biochem Biophys Res Commun 2004; 324(3): 986-992.
REFERENCE LINK - Vogelstein B, Lane D, and Levine AJ.. Stabilization of p53 and transactivation of its target genes in response to replication blockade. Nature 2000; 408: 307-310.
REFERENCE LINK - Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 2010; 108: 73-112.
REFERENCE LINK - Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004; 64(24): 9152-9159.
REFERENCE LINK - Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF et al. Improved ATM kinase inhibitor KU-60019 radiosensitises glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther 2009; 10: 2894-2902.
REFERENCE LINK - Rainey MD, Charlton ME, Stanton RV, Kastan MB. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res 2008; 68(18): 7466-7474.
REFERENCE LINK - Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer 2011; 105(3): 372-381.
REFERENCE LINK - Alao JP, Sunnerhagen P. The ATM and ATR inhibitors CGK733 and caffeine suppress cyclin D1 levels and inhibit cell proliferation. Radiat Oncol 2009; 4: 51.
REFERENCE LINK - Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 1999; 59(17): 4375-4382.
- Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res 2006; 66(10): 5354-5362.
REFERENCE LINK - Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 1994; 269(7): 5241-5248.
- Sarkaria JN, Randal ST, Ericka CB, Amy PK, David EH, Robert TA. Inhibition of Phosphoinositide 3-Kinase Related Kinases by the Radiosensitizing Agent Wortmannin. Cancer Res 1998; 58: 4375-4382.
- Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther 2008; 7(9): 2955-2966.
REFERENCE LINK - Blasina A, Hallin J, Chen E, Arango ME, Kraynov E, Register J et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther 2008; 7(8): 2394-2404.
REFERENCE LINK - Syljuåsen RG, Sørensen CS, Nylandsted J, Lukas C, Lukas J, Bartek J. Inhibition of Chk1 by CEP-3891 accelerates mitotic nuclear fragmentation in response to ionizing Radiation. Cancer Res 2004; 64(24): 9035-9040.
REFERENCE LINK - McNeely SC, Burke TF, DurlandBusbice SS, Barnard DS, Marshall MS, Bence AK et al. LY2606368, a second generation Chk1 inhibitor, inhibits growth of ovarian carcinoma xenografts either as monotherapy or in combination with standard-of-care agents. Molecular Cancer Therapeutics 2011; 10(11): A108.
- Arienti KL, Brunmark A, Axe FU, McClure K, Lee A, Blevitt J et al.. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem 2005; 48(6): 1873-1885.
REFERENCE LINK - Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene 2001; 20: 7453-7463.
REFERENCE LINK - Riesterer O, Matsumoto F, Wang L, Pickett J, Molkentine D, Giri U et al. A novel Chk inhibitor, XL-844, increases human cancer cell radiosensitivity through promotion of mitotic catastrophe. Invest New Drugs 2011; 29(3): 514-522.
REFERENCE LINK - Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res 2005; 33(15): 4711-4724.
REFERENCE LINK - Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361(2): 123-134.
REFERENCE LINK - Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer 2004; 4(4): 296-307.
REFERENCE LINK - Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res 2008; 14(23): 7917-7923.
REFERENCE LINK - Guha C, Guha U, Tribius S, Alfieri A, Casper D, Chakravarty P et al. Antisense ATM gene therapy: a strategy to increase the radiosensitivity of human tumors. Gene Ther 2000; 7(10): 852-858.
REFERENCE LINK - Collis SJ, Swartz MJ, Nelson WG, DeWeese TL. Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res 2003; 63(7): 1550-1554.
- Truman JP, Gueven N, Lavin M, Leibel S, Kolesnick R, Fuks Z et al. Down-regulation of ATM protein sensitises human prostate cancer cells to radiation-induced apoptosis. J Biol Chem 2005; 280(24): 23262-23272.
REFERENCE LINK - Jung M, Dritschilo A. NF-kappa B signalling pathway as a target for human tumor radiosensitization. Semin Radiat Oncol 2001; 11(4): 346-351.
REFERENCE LINK - te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res 2002; 62(6): 1876-1883.
- Crescenzi E, Palumbo G, de Boer J, Brady HJ. Ataxia telangiectasia mutated and p21CIP1 modulate cell survival of drug-induced senescent tumor cells: implications for chemotherapy. Clin Cancer Res 2008; 14(6): 1877-1887.
REFERENCE LINK - Zou J, Qiao X, Ye H, Zhang Y, Xian J, Zhao H et al. Inhibition ofataxia-telangiectasia mutated by antisense oligonucleotide nanoparticles induces radiosensitization of head and neck squamous-cell carcinoma in mice. Cancer Biother Radiopharm 2009; 24(3): 339-346.
REFERENCE LINK - Neijenhuis S, Verwijs-Janssen M, van den Broek LJ, Begg AC, Vens C. Targeted radiosensitization of cells expressing truncated DNA polymerase {beta}. Cancer Res 2010; 70(21): 8706-8714.
REFERENCE LINK - Golding SE, Rosenberg E, Adams BR, Wignarajah S, Beckta JM, O’Connor MJ et al. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle 2012; 11(6): 1167-1173.
- Roossink F, Wieringa HW, Noordhuis MG, Ten Hoor KA, Kok M, Slagter-Menkema L et al. The role of ATM and 53BP1 as predictive markers in cervical cancer. Int J Cancer 2012; 131(9): 2056-2066.
REFERENCE LINK - Zhelev NZ, Todorov IT, Philipova RN, Hadjiolov AA. Phosphorylation-related accumulation of the 125K nuclear matrix protein mitotin in human mitotic cells. J Cell Sci 1990; 95: 59-64.
- Zhang GJ, Safran M, Wei W, Sorensen E, Lassota P, Zhelev N et al. Bioluminescent imaging of Cdk2 inhibition in vivo. Nat Med 2004; 10(6): 643-648.
REFERENCE LINK - Muslimovic A, Ismail IH, Gao Y, Hammarsten O. An optimized method for measurement of gamma-H2AX in blood mononuclear and cultured cells. Nat Protoc 2008; 3(7): 1187-1193.
REFERENCE LINK - Johnson SA, You Z, Hunter T. Monitoring ATM kinase activity in living cells. DNA Repair (Amst) 2007; 6(9): 1277-1284.
REFERENCE LINK - Bin Zhang, Xiubin Gu, Uppalapati U, Ashwell MA, Leggett DS, Li CJ. High-content fluorescent-based assay for screening activators of DNA damage checkpoint pathways. J Biomol Screen 2008; 13(6):538-543.
REFERENCE LINK - Tummala H, Khalil HS, Nikolova E, Mitev V, Zhelev N. Cell-based nanosensors for systems biology research and drug development. Curr Opin Biotechnol 2011; 22(1) S25.
REFERENCE LINK - Walker PR, Carson C, Leblanc J, Sikorska M. Labeling DNA damage with terminal transferase. Applicability, specificity, and limitations. Methods Mol Biol 2002; 203: 3-19.
- Chesselet MF, MacKenzie L, Hoang T. Detection of DNA damage in tissue sections by in situ nick translation. Curr Protoc Neurosci 2001; Chapter 1: Unit 1.9.
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 1988; 175(1): 184-191.
REFERENCE LINK - Fernández JL, Goyanes VJ, Ramiro-Díaz J, Gosálvez J. Application of FISH for in situ detection and quantification of DNA breakage. Cytogenet Cell Genet 1998; 82(3-4): 251-256.
REFERENCE LINK



